Session Information
Date: Tuesday, November 14, 2023
Title: (2095–2140) RA – Diagnosis, Manifestations, and Outcomes Poster III
Session Type: Poster Session C
Session Time: 9:00AM-11:00AM
Background/Purpose: Upadacitinib (UPA), an oral Janus kinase (JAK) inhibitor, significantly improves the signs and symptoms of RA patients1-3. The present study aims to elucidate the mechanism of action of UPA by integrating changes in a genome-wide analysis of methylome data and the blood transcriptome over time from patients enrolled in three phase-3 studies of UPA.
Methods: Longitudinal (baseline [BL], week [W] 2 and W12]) peripheral whole blood samples were randomly selected for analysis (SELECT-NEXT [1]; placebo [PBO], n = 62; UPA 15 mg once daily [QD], n = 61), SELECT-BEYOND [2] (PBO, n = 53; UPA 15 mg QD, n = 55) and SELECT-COMPARE [3] (PBO, n = 181; UPA 15 mg QD, n = 101). This analysis assessed the impact of UPA treatment on all subjects regardless of change in the disease activity. The biospecimen were used to analyze gene expression with a transcriptome-wide gene-level expression profiling tool and methylation profiling with a genome-wide methylation screening tool array. Transcriptome data from the 3 studies were combined following single channel array normalization. Linear models for microarray and RNA-Seq data model (LIMMA package) were used to evaluate transcriptome and methylation changes at W2 and W12 compared to BL. The threshold of significance was defined by false discovery rate < 0.05 and log2 fold-change above the levels observed in the PBO samples. Canonical and Upstream Regulators Pathway analyses were performed with ingenuity pathway analysis (IPA).
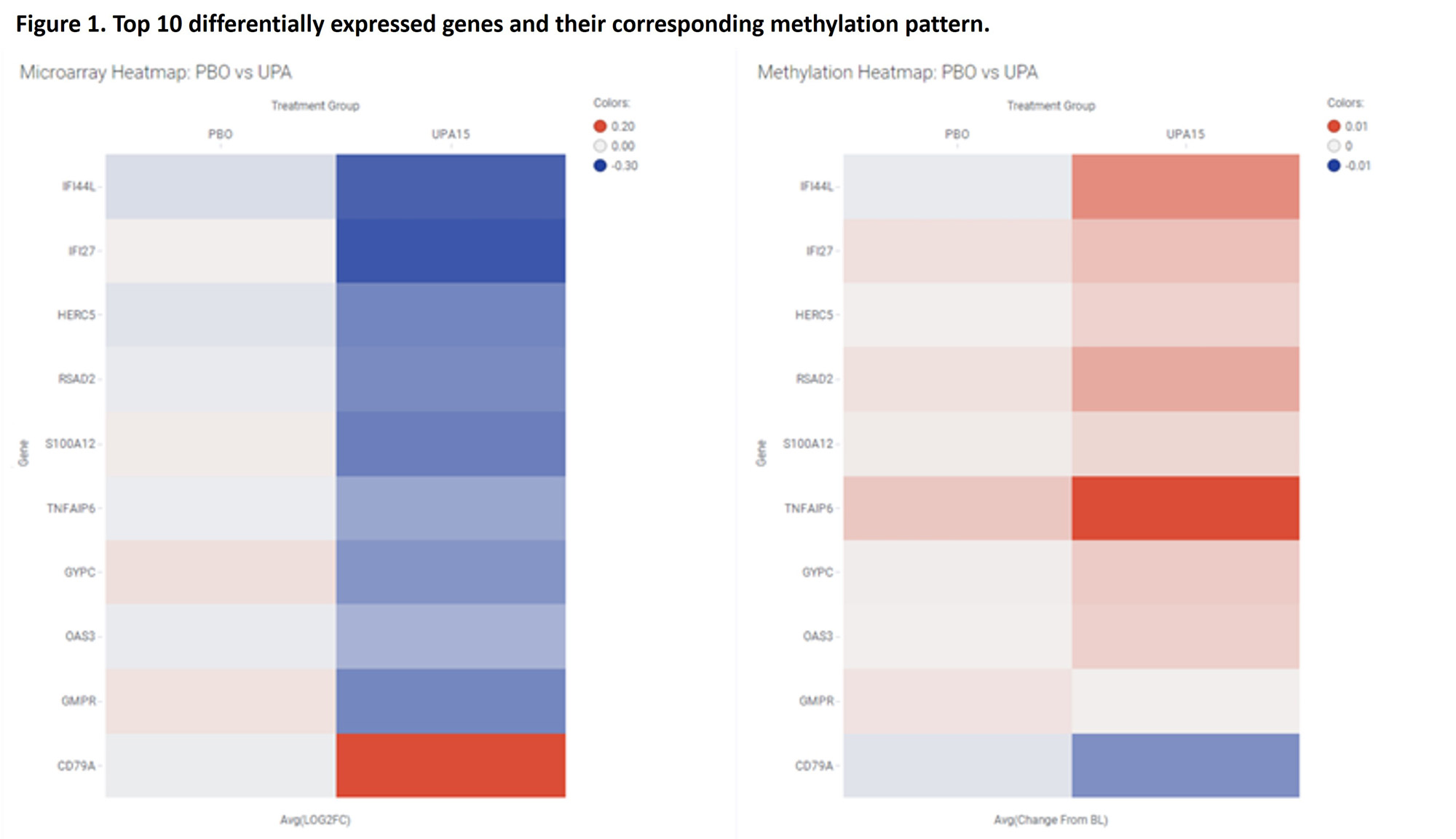
Results: The transcripts for 294 genes were differentially expressed (DE) at W2, of which 114 remained DE at W12. An additional 98 genes were DE at W12 vs baseline. Pathway analysis of DE genes shows early modulation of key pathways like IFN, IL10 and S100 signaling. Furthermore, pathway analysis predicted that UPA downregulates pathways related to IL1B, TNF, types 1 and 2 IFN, IL17A, and IL6 biology. The methylation data covered 282 genes out of the 294 identified above. Of the 282, 34% (n=97) of the genes were significantly differentially methylated and consistent with the RNA expression data. Pathway analysis based on these 97 genes predicted the upstream downregulation of inflammatory regulators such as TNF, IFNα, TGF-β, IL1B, and IL17.
Conclusion: The present study reveals the effect of treatment with UPA on the transcription machinery of leukocytes from RA patients at the RNA and DNA methylation levels. While UPA may directly affect the methylation state and expression of target genes, we cannot exclude that some of the observed changes reflect a change in blood cell composition. This integrated analysis confirms that UPA normalizes key pathological pathways associated with RA, expanding our understanding of its mode of action in RA and creates a framework for multi-omic biomarker discovery for response to JAK inhibitors.
1. Burmester GR, et al. (2018) Lancet 391:2503–12.
2. Genovese MC, et al. (2018) Lancet 391:2513–24.
3. Fleischmann R, et al. (2019) Arthritis Rheumatol 71:1788–800
To cite this abstract in AMA style:
Lal P, Xu Y, Sornasse T, Sharma R, Jafarpour L, Camp H, McInnes I. Integrated Analysis of Gene Expression and Methylation Identifies Biomarkers Associated with Mode of Action of Upadacitinib Treatment in Rheumatoid Arthritis [abstract]. Arthritis Rheumatol. 2023; 75 (suppl 9). https://acrabstracts.org/abstract/integrated-analysis-of-gene-expression-and-methylation-identifies-biomarkers-associated-with-mode-of-action-of-upadacitinib-treatment-in-rheumatoid-arthritis/. Accessed .« Back to ACR Convergence 2023
ACR Meeting Abstracts - https://acrabstracts.org/abstract/integrated-analysis-of-gene-expression-and-methylation-identifies-biomarkers-associated-with-mode-of-action-of-upadacitinib-treatment-in-rheumatoid-arthritis/