Session Information
Date: Sunday, October 26, 2025
Title: (0067–0097) Rheumatoid Arthritis – Etiology and Pathogenesis Poster
Session Type: Poster Session A
Session Time: 10:30AM-12:30PM
Background/Purpose: The mucosal origins hypothesis of RA proposes that immune responses to microorganisms at mucosal sites (e.g. intestine) lead to systemic inflammation and autoimmunity. Little is known about how the fecal microbiome composition and functional capacity may evolve in the pre-clinical period to contribute to development of clinical inflammatory arthritis (IA). The study objective was to compare the fecal microbiome over time among individuals at-risk for RA (ARI) who converted to IA (Converters) versus those who did not (Non-Converters).
Methods: Fecal samples for shotgun metagenomic sequencing of fecal DNA by Illumina NovaSeqX were prospectively collected intermittently over 3 years from ARI, defined as adults with serum cyclic citrullinated peptide antibodies (CCP3 ³20) with no IA at baseline. ARI were grouped into Converters, who developed RA-like synovitis of ³1 joint during follow-up, and Non-Converters. Taxonomic and functional microbiome profiling was performed by MetaPhlAn4 and HUMAnN3. Differential abundance was assessed by MaAsLin3 by mixed linear models. A single baseline and follow up sample from each participant were analyzed. Follow up samples selected were those obtained at the timepoint closest to identification of IA in Converters and the timepoint in Non-Converters that was closest to the median number of days after baseline that Converter follow ups were obtained.
Results: There were no significant differences between Converters (n=14) and Non-Converters (n=31) in baseline demographics and labs (Table 1). Taxonomic analysis demonstrated -7.5 log2fold lower Agathobaculum butyriciproducens t_14991 in Converters compared to Non-Converters at baseline; multiple other taxa were differentially abundant but not statistically significant after false discovery rate (FDR) (Figure 1). Between baseline and follow up, Converters had a greater increase in A. butyriciproducens t_14991, Pseudoruminococcus massiliensis, and Adlercreutzia equolifaciens; and a greater decrease in Turicibacter sanguinis; not significant after FDR. Functional analysis showed significant alterations in fatty acid degradation pathways at baseline among Converters (Figure 2). Longitudinally, fatty acid degradation gene features and pathways in Converters changed more significantly over time.
Conclusion: Among a longitudinal cohort of ARI, Converters showed baseline depletion of immunoregulatory bacteria A. butyriciproducens t_14991, a short-chain fatty acid (SCFA) producer. Moreover, the trend of changes in several SCFA-producing and starch-metabolizing bacteria in Converters at baseline and over time suggest that Converters harbor gut microbiomes with altered inflammation-regulating capacity compared to Non-Converters. Many of the predicted functional changes at baseline and over time in Converters involve metabolism of immunomodulatory metabolites, including fatty acid degradation pathways. Together, these data indicate complex feedback interactions between the gut microbiome and the host during IA development. Further exploration into longitudinal gut microbiome changes in ARI is needed to better understand potential diagnostic, preventative, and therapeutic applications.
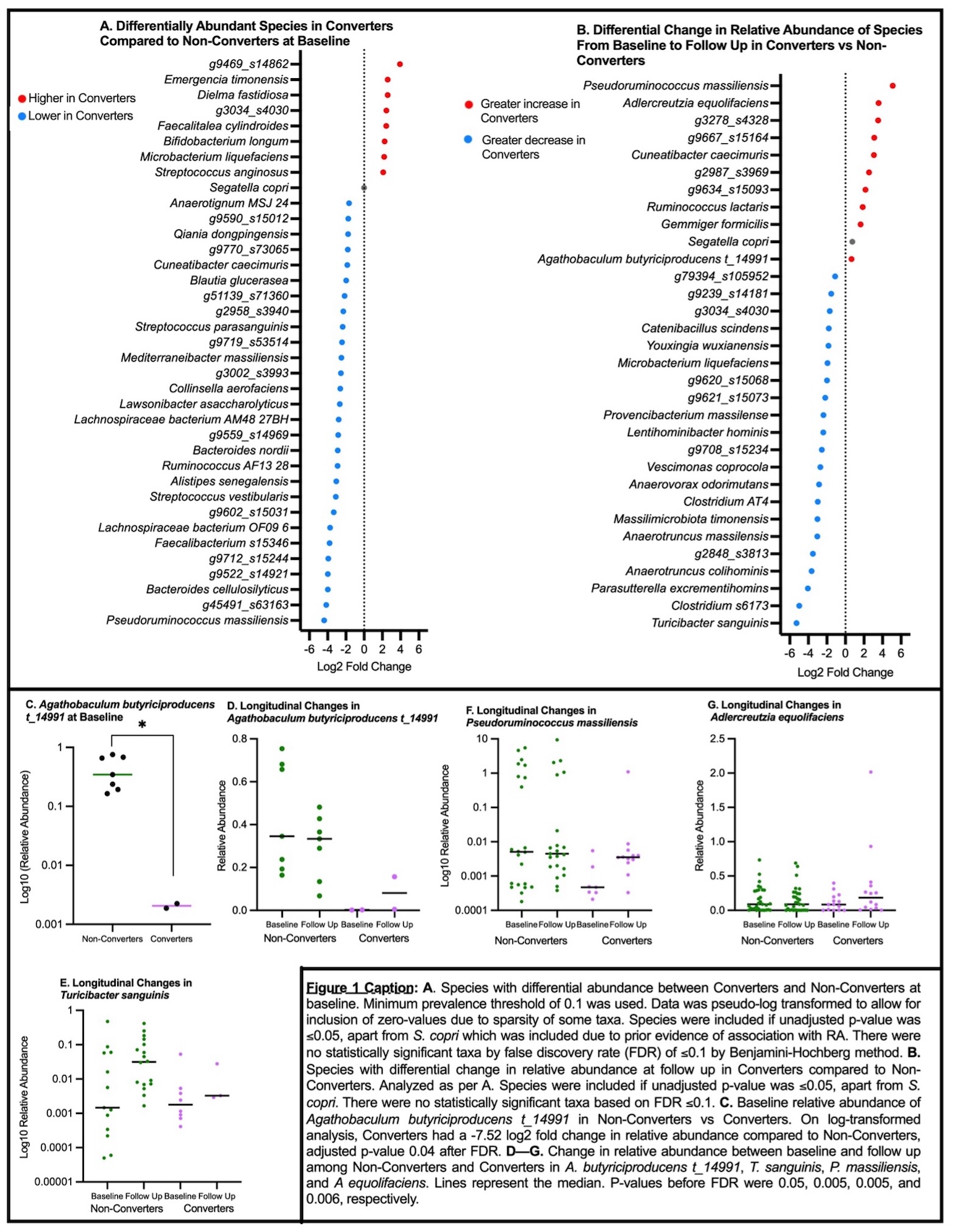 Table 1: Participant Characteristics and Laboratory Values
Table 1: Participant Characteristics and Laboratory Values
.jpg) Figure 1: Taxonomic Differences Between Converters and Non-Converters at Baseline and Follow Up
Figure 1: Taxonomic Differences Between Converters and Non-Converters at Baseline and Follow Up
.jpg) Figure 2: Predicted Functional Gene Features and Pathway Differences Between Converters and Non-Converters at Baseline and Follow Up
Figure 2: Predicted Functional Gene Features and Pathway Differences Between Converters and Non-Converters at Baseline and Follow Up
To cite this abstract in AMA style:
Cole L, Allen B, Liu S, Feser M, Phyo L, Moss L, Frank D, Harris J, Demoruelle K, Deane K, Holers V, Kuhn K. Imbalance of Inflammation-Regulating Microorganisms and Predicted Metabolomic Pathways Associates With Disease Evolution in Individuals At-Risk for Rheumatoid Arthritis [abstract]. Arthritis Rheumatol. 2025; 77 (suppl 9). https://acrabstracts.org/abstract/imbalance-of-inflammation-regulating-microorganisms-and-predicted-metabolomic-pathways-associates-with-disease-evolution-in-individuals-at-risk-for-rheumatoid-arthritis/. Accessed .« Back to ACR Convergence 2025
ACR Meeting Abstracts - https://acrabstracts.org/abstract/imbalance-of-inflammation-regulating-microorganisms-and-predicted-metabolomic-pathways-associates-with-disease-evolution-in-individuals-at-risk-for-rheumatoid-arthritis/
