Session Information
Session Type: Poster Session (Tuesday)
Session Time: 9:00AM-11:00AM
Background/Purpose: Given the heterogeneity of systemic lupus erythematosus (SLE), the effect of any intervention is expected to vary. The ability to identify those most and least likely to benefit from a treatment would improve the interpretability of trial outcomes and advance medical care. Conventional subgroup analyses suffer from low power, can encompass only a few variables at a time, and require a priori specification of cut-points for continuous variables. We explored the utility of a machine learning-based algorithm for discovering in a SLE trial the subgroups in which adding experimental therapy to standard of care considerably enhances or diminishes response compared to placebo (PBO).
Methods: A two-step “virtual twin” (VT) method was applied to combined data from the BLISS-52 (N=865) and BLISS-76 (N=819) trials. A random forest algorithm was first used to estimate for each patient, given baseline characteristics, the probabilities of SRI-4 response to belimumab and PBO. A regression tree was then constructed to partition the study population into distinct subgroups and identify those in which the estimated difference in these response probabilities is much greater or smaller than the treatment effect in the overall population. Two separate VT analyses were conducted of the 10 mg/kg and 1 mg/kg belimumab doses compared to PBO. Cross-validation was used to assess the method’s performance.
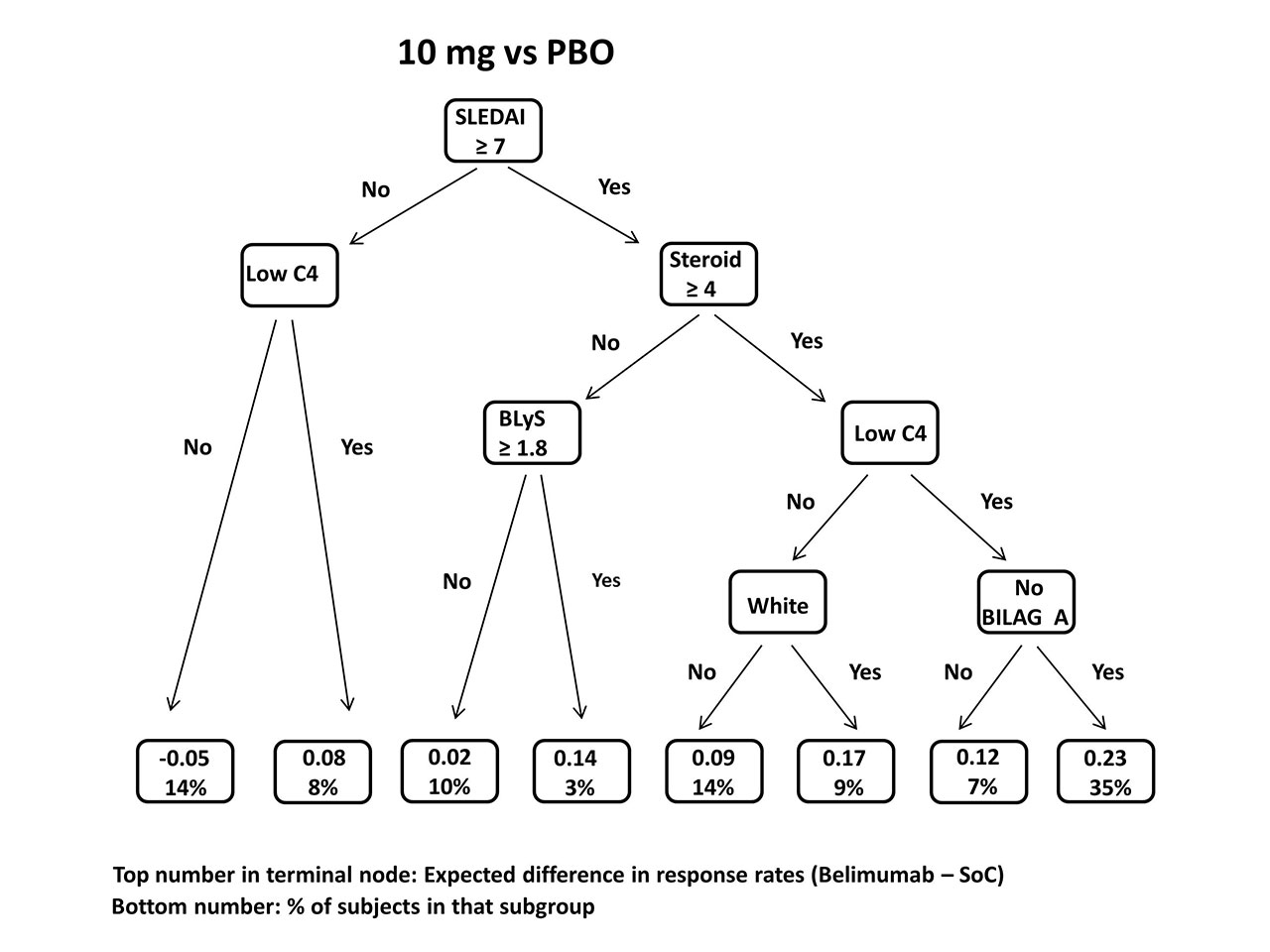
Results: In the combined BLISS trials, response rates to the primary endpoint (SRI-4) were 51% in those receiving 10 mg/kg belimumab, 46% (1 mg/kg), and 39% (PBO). VT analysis of 10 mg/kg vs. PBO found a 23% belimumab response advantage over PBO in patients with SLEDAI ≥ 7 & steroid dose ≥ 4 mg/d & low C4 & no BILAG A at baseline, vs 12% in the total population. In contrast, the estimated response difference in those entering with SLEDAI < 7 & normal C4 was 5% lower on 10 mg/kg than PBO. In analysis of 1 mg/kg vs. PBO, two subgroups showed enhanced belimumab effect: SLEDAI ≥ 8 & steroid dose ≥ 19 mg/d and SLEDAI ≥ 8 & steroid dose < 19 mg/d & BLyS ≥ 1.9 ng/mL; average estimated between-treatment response differences were 18% and 14%, respectively, compared to 7% in the overall population. But in patients with SLEDAI < 8 & steroid dose < 16 mg/d & age < 43, the 1 mg/kg belimumab response rate was estimated to be 7% lower. Cross-validation indicated the accuracy of the VT method to identify subgroups exceeded 70%.
Conclusion: Enhanced belimumab response was associated with low C4 and higher disease activity, steroid dose, and BLyS levels, as in prior studies. However, the VT method identified alternative cutpoints for continuous variables and additional features predicting non-response. SLEDAI ≥ 7 or 8 was most predictive of response to treatment. Thus, lower response difference is identified in patients who are potentially too ill (BILAG A severity) or not ill enough (minimal disease criteria) to benefit from adding belimumab. The 1 mg/kg belimumab effect was enhanced only in those on high baseline steroid doses. The VT and other machine learning techniques are promising for subgroup discovery in SLE trials as more sophisticated biomarkers, especially potent but less common indicators, become available.
To cite this abstract in AMA style:
Kim M, Pradhan K, Izmirly P, Kalunian K, Hanrahan L, Merrill J. Identifying Subgroups of SLE Patients with Differential Responses to a BLyS Inhibitor: Application of a Machine Learning Algorithm to Clinical Trial Data [abstract]. Arthritis Rheumatol. 2019; 71 (suppl 10). https://acrabstracts.org/abstract/identifying-subgroups-of-sle-patients-with-differential-responses-to-a-blys-inhibitor-application-of-a-machine-learning-algorithm-to-clinical-trial-data/. Accessed .« Back to 2019 ACR/ARP Annual Meeting
ACR Meeting Abstracts - https://acrabstracts.org/abstract/identifying-subgroups-of-sle-patients-with-differential-responses-to-a-blys-inhibitor-application-of-a-machine-learning-algorithm-to-clinical-trial-data/