Session Information
Session Type: Poster Session B
Session Time: 9:00AM-10:30AM
Background/Purpose: One of the hallmarks of SLE is clinical and molecular heterogeneity. Recent work has leveraged next-generation sequencing techniques to identify molecular signatures characteristic of subsets of patients’ disease. Here, we employed gene set variation analysis (GSVA) of informative gene modules coupled with machine learning techniques to identify molecular endotypes of SLE patients based on dysregulation of specific biologic pathways, and interrogated them for clinical utility.
Methods: Gene expression profiles from 1620 active, female patients fulfilling ≥4 1997 ACR criteria for the diagnosis of SLE (GSE88884) were analyzed by GSVA. 32 molecular signatures relating to immune cells and inflammatory processes that distinguish active SLE from inactive SLE and/or non-lupus healthy controls were used to query the data and GSVA enrichment scores for each of the 32 features were assigned to individual patients. Iterative k-means clustering optimized by the sum of squared distances of samples to their closest cluster center stratified patients by feature enrichment into six clusters, determined to be optimal by the elbow and Silhouette methods. Training, validation, and testing of unrelated datasets were carried out using multiple machine learning algorithms. The six endotypes were finally characterized by associated patient metadata and response to the anti-BAFF therapy, tabalumab.
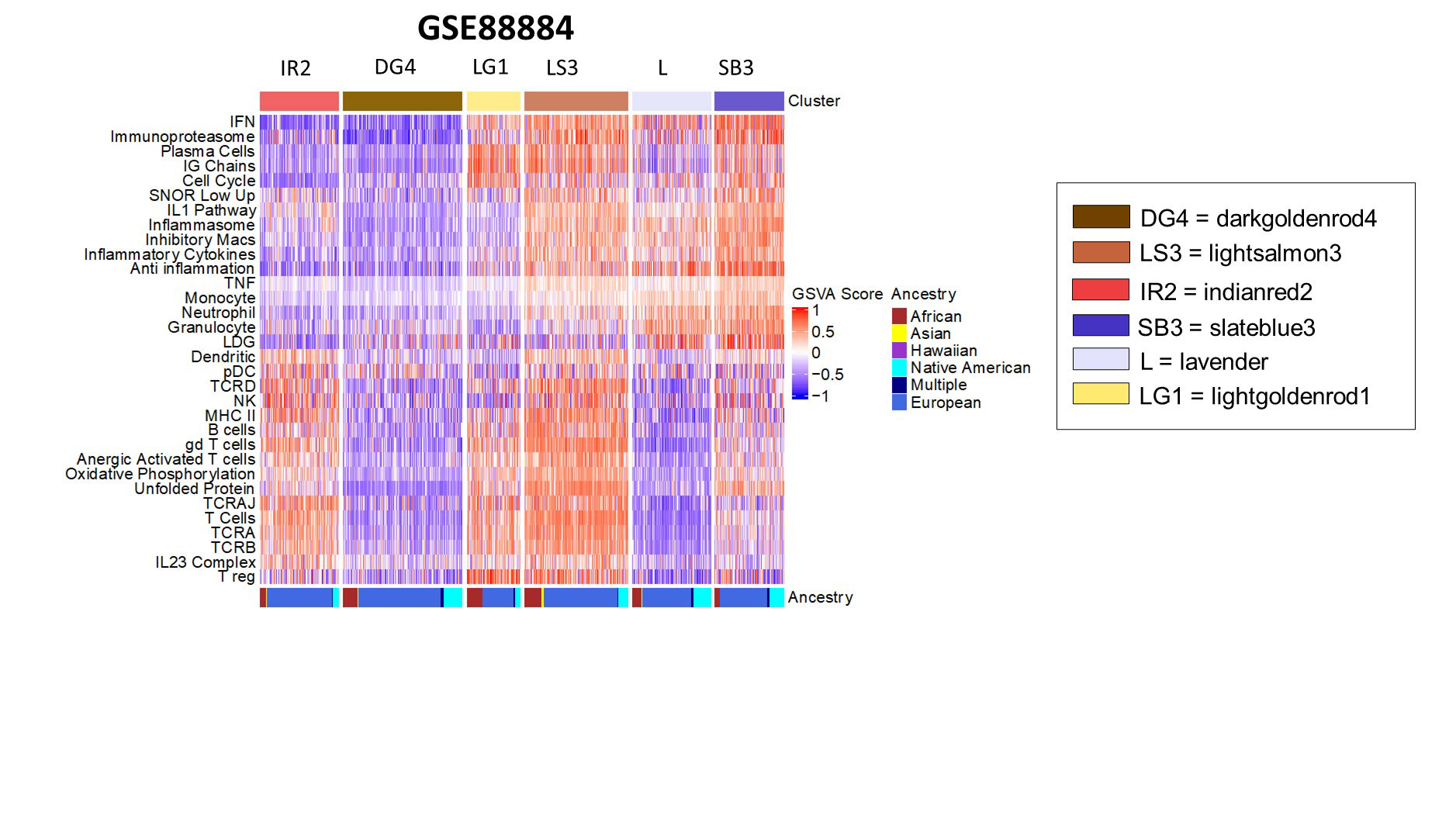
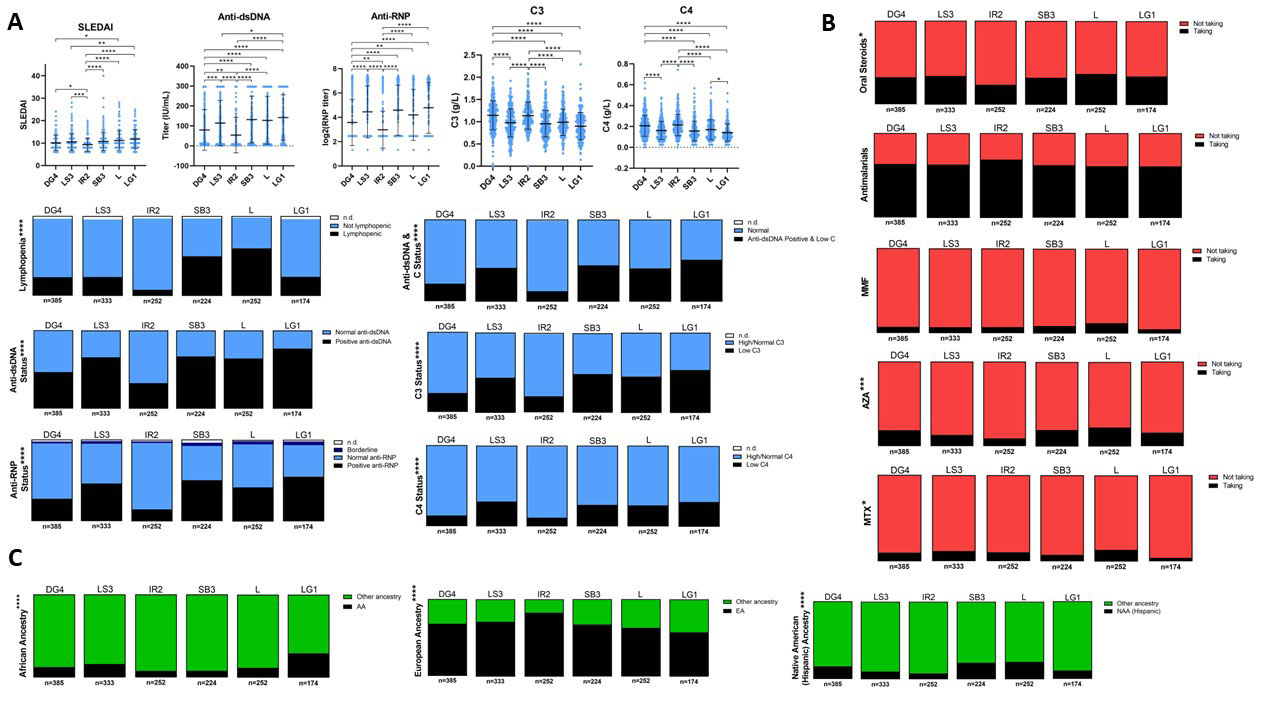
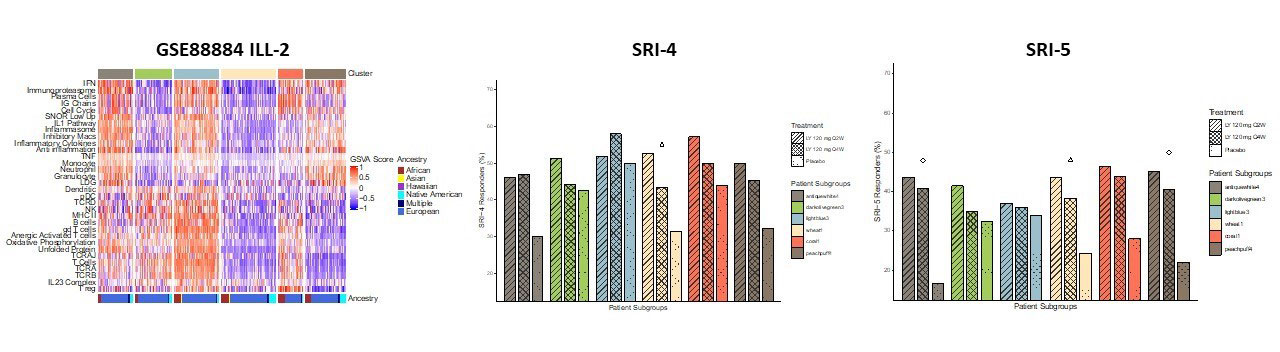
Results: Gene expression profiles of adult SLE patients by 32 immune and inflammatory features revealed six distinct endotypes (Figure 1). Machine learning algorithms demonstrated a sensitivity of 97% and a specificity of 99% in predicting subset membership and cross-validation with unrelated datasets revealed excellent model performance with a positive predictive value of 96%. Characterization by associated patient metadata revealed significant differences among the endotypes by disease activity, with the subset having the least abnormal profile manifesting the lowest SLEDAI, lowest ANA titers, highest serum complement levels, and lowest incidence of lymphopenia, and the subsets with more abnormal transcriptional profiles showing significantly more abnormal clinical features (Figure 2). There were also significant differences among the endotypes in the use of corticosteroids and immunosuppressive agents. In addition, the endotype with the least abnormal transcriptional profile had a significantly lower frequency of flares over the subsequent 52 weeks (OR=0.116, p=0.00041) and no significant response to the investigational product product used in the trial (tabalumab, Figure 3).
Conclusion: Transcriptomic profiling using GSVA and machine learning identified molecular endotypes of lupus patients with significant differences in clinical outcomes and responsiveness to therapy.
Iterative k-means clustering of GSVA scores of the 32 features in 1620 adult lupus patients from GSE88884 yielded six clusters using baseline gene expression. Shorthand labels indicate patient clusters and color names were randomly generated using the ‘grDevices’ color palette in R. Native American ancestry refers to Hispanic patients.
Quantitative and categorical metadata were summarized for each cluster using baseline values. Metadata was categorized by (A) immunologic/inflammatory and systemic disease indicators, (B) medication use, and (C) patient ancestry. Shorthand labels indicate patient clusters and color names were randomly generated using the ‘grDevices’ color palette in R. Scatterplots in (A) display the mean±SD for each cluster; statistical differences were found with Dunn’s multiple comparisons test. Lymphopenia was defined as less than 1 billion lymphocytes per liter. Significant associations between categorical variables and endotype cluster (denoted with asterisks) were identified using Chi Square test of independence. Missing data (n.d.) were excluded from analyses. Graphs were created in GraphPad Prism v 9.1.00(221). ESR = erythrocyte sedimentation rate. HCQ = hydroxychloroquine. MMF = mycophenolate mofetil. AZA = azathioprine. MTX = methotrexate. AA = African ancestry. EA = European ancestry. NAA = Native American ancestry (refers to Hispanic populations).
*p<0.05; **p<0.01; ***p<0.001; ****p<0.0001
K-means clustering of Illuminate_2 lupus samples and their clinical responses by SRI_4 and SRI-5 per endotype determined by gene expression data and 32 immune/inflammatory features. Responses among the treatment groups were ascertained by the Trend Chi Square test. Endotype color labels were randomly generated using the ‘grDevices’ color palette in R. Q2W indicates frequency of drug administration was every 2 weeks. Q4W indicates frequency of drug administration was every 4 weeks.
∆: p<0.05 observed by Trend Chi Square Test for Q2W>Q4W>Placebo, Q2W>Placebo, and Q2W+Q4W>Placebo.
□: p<0.05 observed by Trend Chi Square Test for Q4W>Placebo and Q2W+Q4W>Placebo
◊: p<0.05 observed by Trend Chi Square Test for Q2W>Q4W>Placebo, Q2W>Placebo, Q4W>Placebo, Q2W+Q4W>Placebo
To cite this abstract in AMA style:
Hubbard E, Bachali P, Kingsmore Allison K, He Y, Catalina M, Grammer A, Lipsky P. Identification of Systemic Lupus Erythematosus Endotypes by Analysis of Inflammatory and Immunologic Gene Expression Signatures [abstract]. Arthritis Rheumatol. 2022; 74 (suppl 9). https://acrabstracts.org/abstract/identification-of-systemic-lupus-erythematosus-endotypes-by-analysis-of-inflammatory-and-immunologic-gene-expression-signatures/. Accessed .« Back to ACR Convergence 2022
ACR Meeting Abstracts - https://acrabstracts.org/abstract/identification-of-systemic-lupus-erythematosus-endotypes-by-analysis-of-inflammatory-and-immunologic-gene-expression-signatures/