Session Information
Date: Sunday, November 13, 2022
Title: RA – Treatment Poster II
Session Type: Poster Session B
Session Time: 9:00AM-10:30AM
Background/Purpose: MTX is the recommended first treatment for rheumatoid arthritis (RA); however, only ~40% respond adequately to MTX. Significant joint damage can occur in the early phase of RA, and response to the first treatment is an indicator of long-term prognosis. Changes in DNA methylation (DNAm) associated with response to these treatments are potential biomarkers for prediction of treatment response.
Methods: We estimated changes in cell-specific DNAm associated with MTX response from whole blood samples collected from RA patients before and after initiation of MTX treatment. Patients included in this study were from the Rheumatoid Arthritis Medication Study (RAMS) and University of California, San Francisco Rheumatoid Arthritis study (UCSF-RA) (Total n=77). All patients met the ACR RA criteria. Blood samples were collected at baseline and after treatment (UCSF-RA: 3-6 months, RAMS: 4 weeks). DAS28-CRP was collected at baseline and after 3-6 months of treatment. Genome-wide methylation profiles were generated with Illumina 450K (RAMS) and EPIC BeadChips (UCSF-RA) from whole blood. Functional normalization and background subtraction with dye-bias normalization and other QC procedures were performed using minfi. Differences between 450K and EPIC platforms were adjusted using Harman. MTX response was defined using the EULAR criteria for DAS28-CRP (Responder: good/moderate response, Non-responder: no response). Differentially methylated positions (DMPs) were identified using limma and Tensor Composition Analysis (TCA). TCA is a method for identifying cell-specific differential DNAm at the CpG level from bulk tissue. B cells, CD4 and CD8 T cells, monocytes, neutrophils, and Natural Killer (NK) cells were included in cell-specific analyses. Linear models evaluated differential DNAm between MTX response groups over time and within each time point. Sex, age, smoking history, estimated global cell-proportions, and batch were included as covariates. Differentially Methylated Regions (DMRs) were identified using Comb-p.
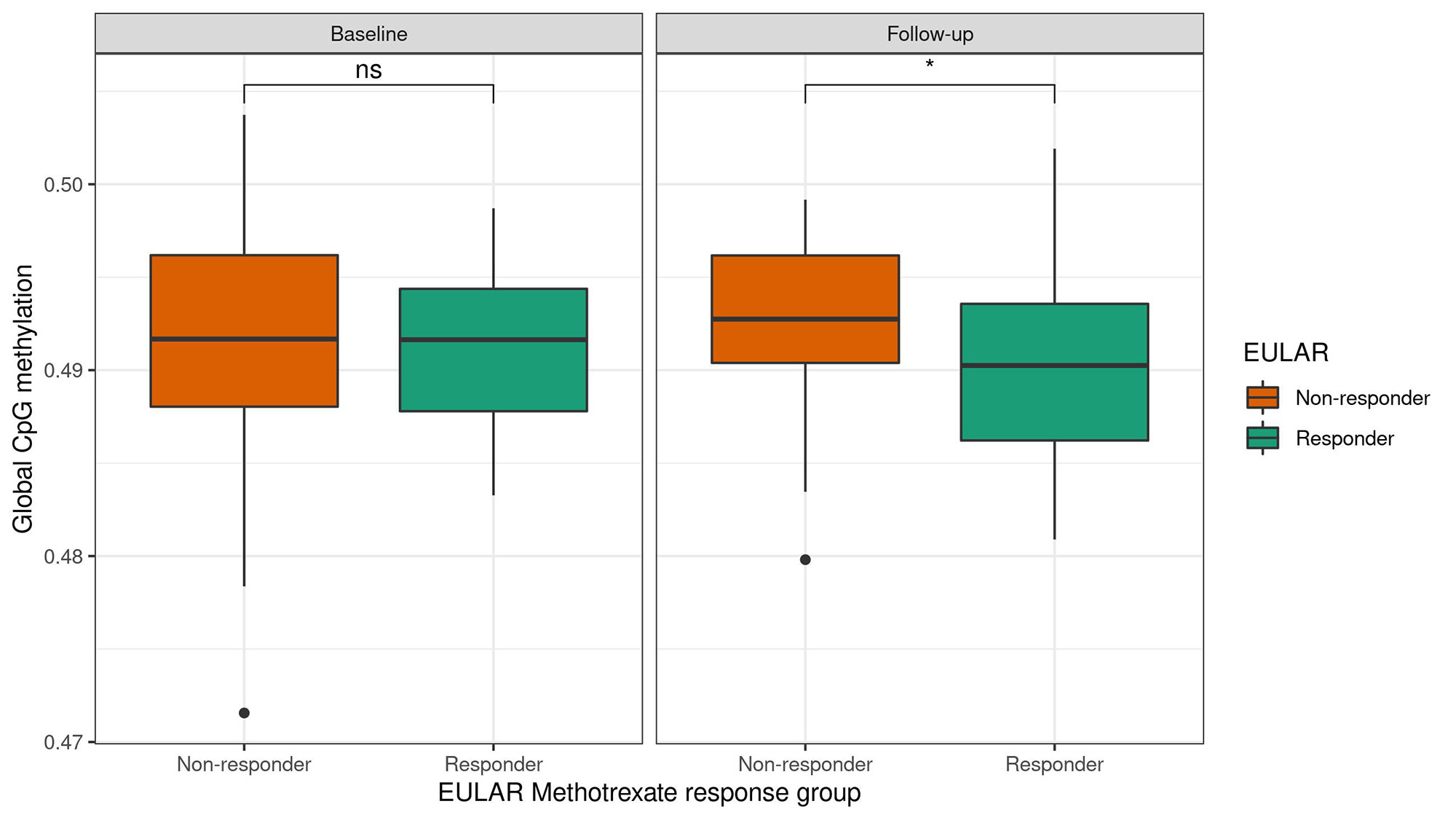
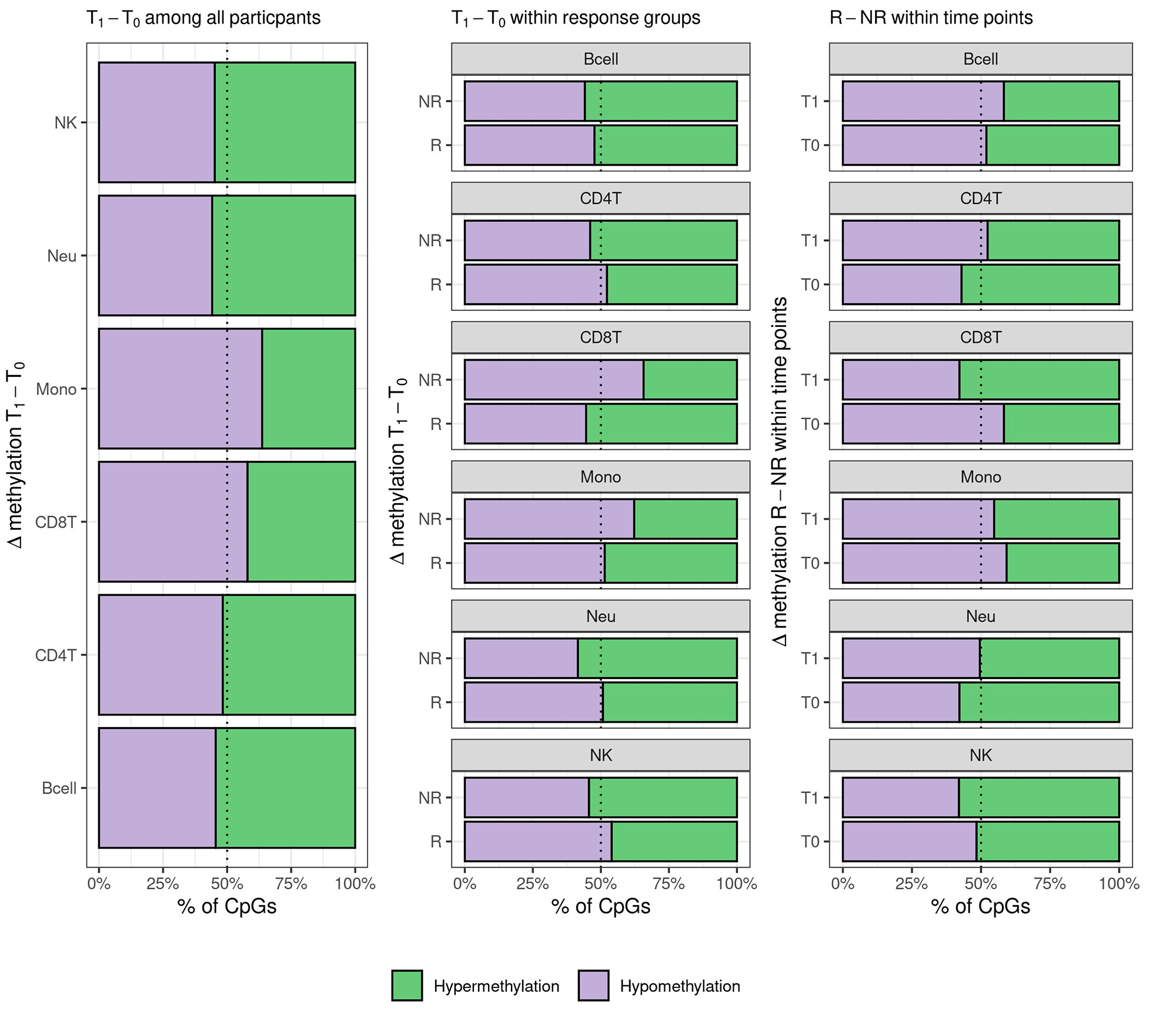
Results: We found evidence for differential global methylation between response groups after treatment (Figure 1). Further, we found patterns of cell-specific differential global methylation associated with MTX response (Figure 2). One DMP was associated at genome wide significance with differential DNAm between responders and non-responders at baseline in CD4T, CD8T, and NK cells (Table 1). Additionally, we identified 39 cell-specific DMRs associated with MTX response. There were no significant findings in whole blood analyses.
Conclusion: We identified cell-specific changes in DNAm associated with MTX response in RA patients. Future studies into DNAm and MTX response should include measurements of DNAm from sorted cells.
To cite this abstract in AMA style:
Adams C, Nair N, Quach H, Quach D, Nititham J, Nakamura M, Graf J, Criswell L, Barcellos L. Identification of Cell-Specific DNA Methylation Changes Associated with MTX Treatment Response in Rheumatoid Arthritis [abstract]. Arthritis Rheumatol. 2022; 74 (suppl 9). https://acrabstracts.org/abstract/identification-of-cell-specific-dna-methylation-changes-associated-with-mtx-treatment-response-in-rheumatoid-arthritis/. Accessed .« Back to ACR Convergence 2022
ACR Meeting Abstracts - https://acrabstracts.org/abstract/identification-of-cell-specific-dna-methylation-changes-associated-with-mtx-treatment-response-in-rheumatoid-arthritis/