Session Information
Date: Monday, November 18, 2024
Title: Systemic Sclerosis & Related Disorders – Basic Science Poster II
Session Type: Poster Session C
Session Time: 10:30AM-12:30PM
Background/Purpose: Systemic Sclerosis (SSc) is a molecularly heterogeneous disease. Distinct subtypes of patients have been identified based on gene expression in skin. In this study, we re-processed genome-wide transcriptomic data of skin biopsies from multiple independent cohorts to generate the largest integrated discovery dataset to date. Semi-supervised clustering was performed to identify SSc intrinsic subtypes and these labels were then used to develop a new robust supervised learning model to predict subtypes in skin.
Methods: Gene expression data from three cohorts (GSE9285, GSE32413, and GSE59787) representing 293 paired forearm and back skin samples from 37 healthy and 137 SSc individuals were processed using a consistent bioinformatic pipeline. Samples were first clustered using constrained k-means, followed by unsupervised k-means clustering on the most heterogeneous group to refine sample groupings. Using the final labels, we developed a set of binary Logistic Regression models with Gene Set Variation Analysis (GSVA) scores to predict Inflammatory, Normal-like, and Fibroproliferative subtypes on new skin samples.
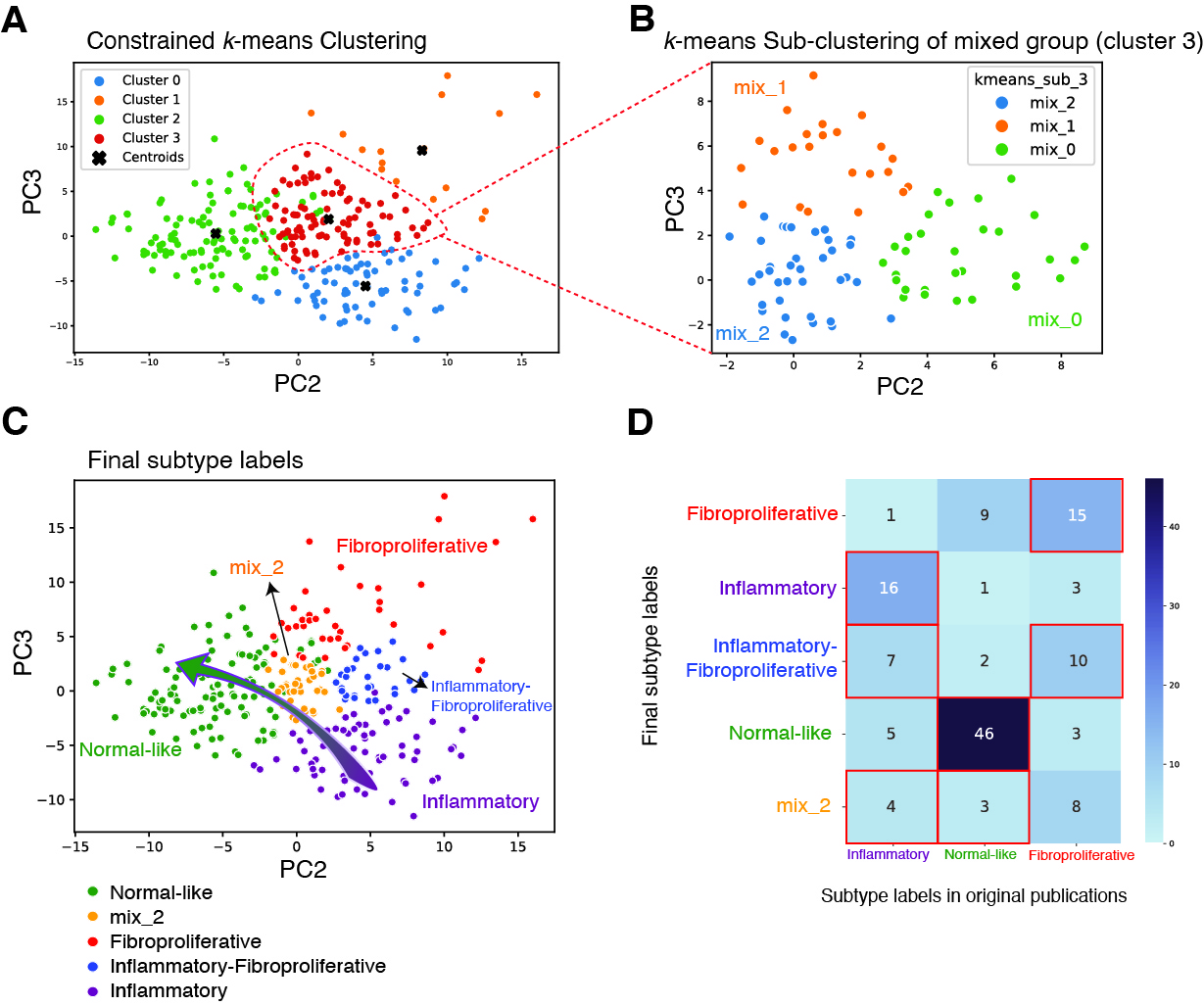
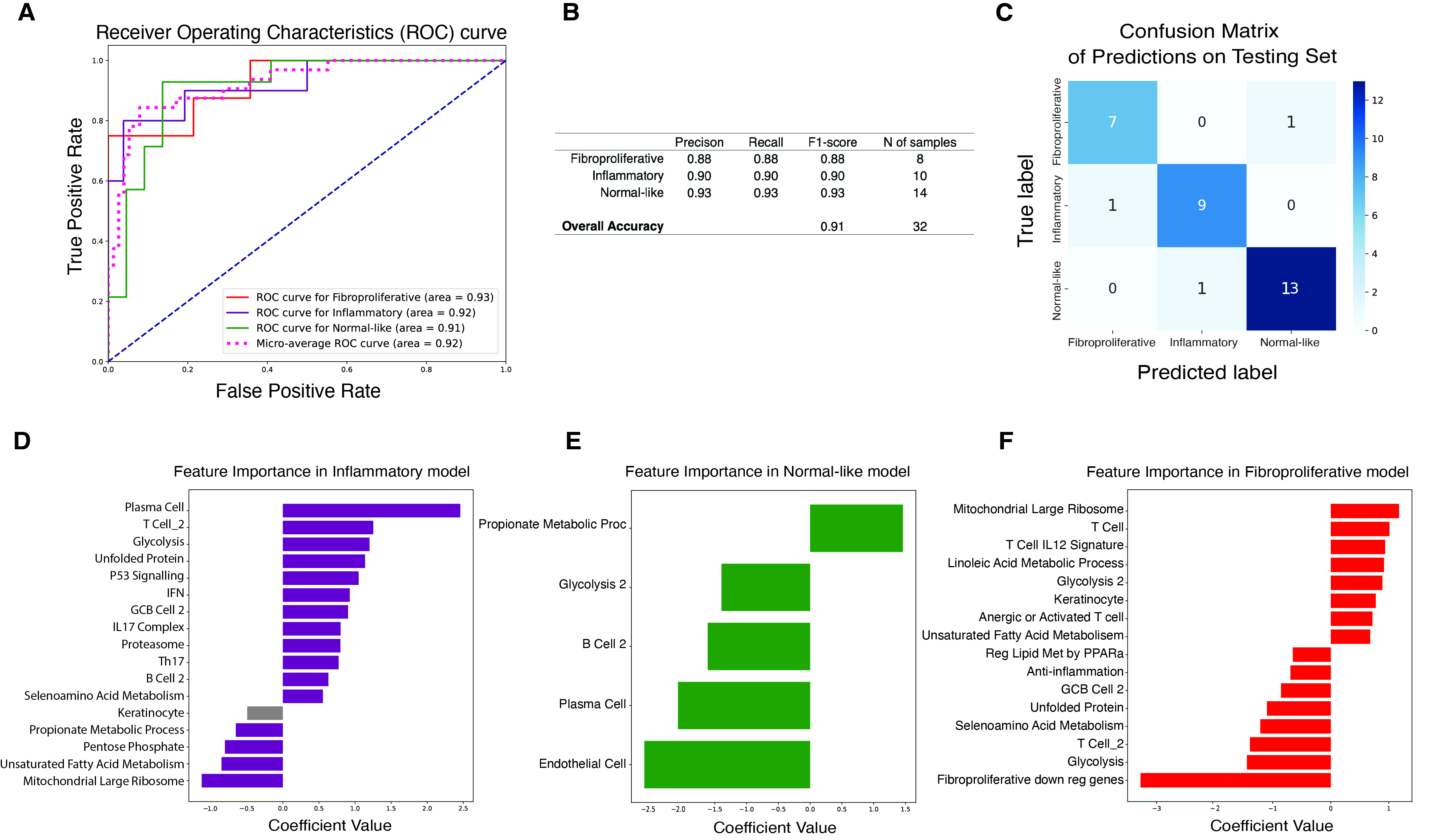
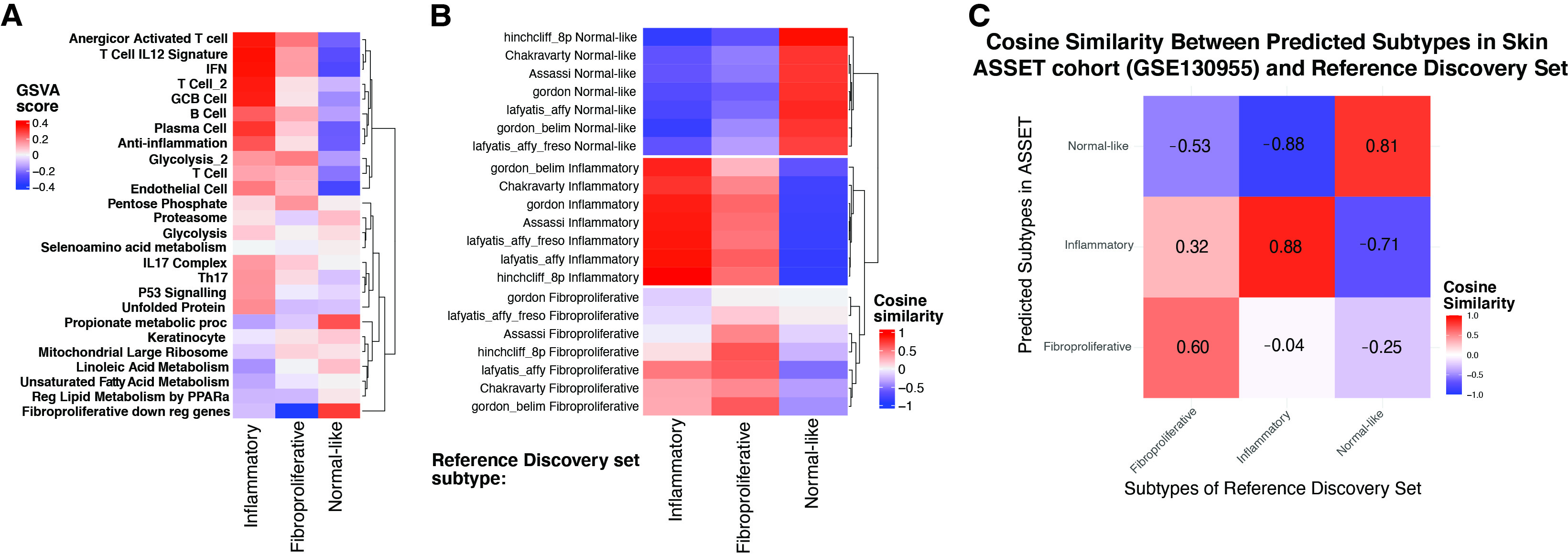
Results: We identified 5 intrinsic molecular subtypes of SSc in this study through semi-supervised clustering and comparison to original publications (Fig. 1A-C). In addition to the previously reported inflammatory, fibroproliferative, and normal-like subtypes, we also found an inflammatory-fibroproliferative group and an intermediate group between inflammatory and normal-like, which may represent a transitional subtype. Analysis across these groups shows that genetic markers of fibrosis, such as COMP and SFRP4, are highest in inflammatory patients and decrease across the transitional group, to its lowest level in normal-like patients. For classification we used the original 3 subtype labels due to limited samples numbers. Data were split with 20% of samples held out for testing, 4-fold cross-validation (CV) was performed with GSVA scores as features for each subtype. The best models were chosen based on test performance, they showed AUROC of 0.92, 0.91, and 0.93 for inflammatory, normal-like, and fibroproliferative subtypes respectively (Figure 2A). The final subtype assignment for each patient was made by taking the highest predicted probability by the three binary models. The final, three-class model shows good precision and recall for each subtype (Figures 2B-C). Additionally, for each subtype we determined the gene sets that are most predictive and their importance (Figures 2D-F). The final model has the ability to identify samples in additional independent cohorts with high similarities to the corresponding reference for each subtype using 16 gene sets in total (Figure 3A-C).
Conclusion: These results extend our previously published subtyping results and support the existence of major gene expression subtypes. Additionally, we identified two intermediate subgroups. The classification model for skin samples, validated across multiple cohorts, shows the robustness of our approach and its potential to enhance patient stratification, leading to personalized treatment in SSc in the future.
To cite this abstract in AMA style:
Gong Z, Parvizi R, Jarnagin H, Chen H, Morrisson M, Wood T, Hinchcliff M, Whitfield M. Identification and Prediction of Systemic Sclerosis Intrinsic Subtypes Using Semi-Supervised and Supervised Learning on Gene Expression Data of Multiple Cohorts [abstract]. Arthritis Rheumatol. 2024; 76 (suppl 9). https://acrabstracts.org/abstract/identification-and-prediction-of-systemic-sclerosis-intrinsic-subtypes-using-semi-supervised-and-supervised-learning-on-gene-expression-data-of-multiple-cohorts/. Accessed .« Back to ACR Convergence 2024
ACR Meeting Abstracts - https://acrabstracts.org/abstract/identification-and-prediction-of-systemic-sclerosis-intrinsic-subtypes-using-semi-supervised-and-supervised-learning-on-gene-expression-data-of-multiple-cohorts/