Session Information
Date: Saturday, November 6, 2021
Title: Abstracts: Vasculitis – Non-ANCA-Associated & Related Disorders (0502–0507)
Session Type: Abstract Session
Session Time: 3:30PM-3:45PM
Background/Purpose: Non-infectious aortitis may be a manifestation of systemic large vessel vasculitis such as giant cell arteritis (GCA) or may be a form of single organ vasculitis termed ‘clinically isolated aortitis’ (CIA). Aortitis is generally diagnosed after aortic aneurysm surgery and histopathologic examination of resected aortic tissue. At current, there is no specific biomarker that assists physicians in their diagnostic and treatment decisions for aortitis. Moreover, a genome-wide expression analysis to detect differences in transcriptomic signatures between inflammatory and non-inflammatory aortic aneurysms has not been conducted to date. To this end, we used global transcriptome profiling (RNA-seq) to provide novel insights into the biomolecular processes underlying inflammatory aortic aneurysms. We also compared the transcriptomic signature of aortic tissue from patients with CIA and GCA.
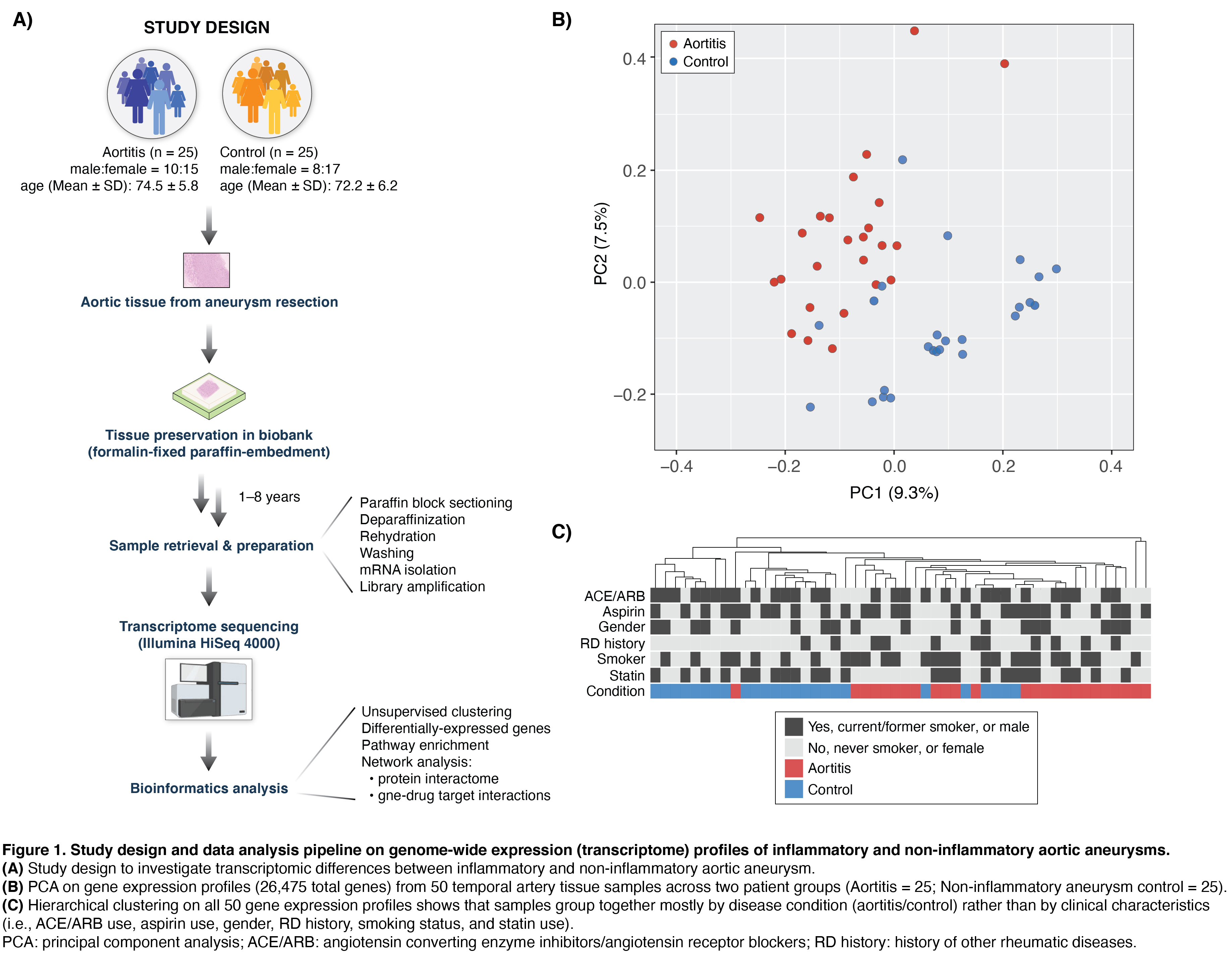
Methods: RNA-seq (HiSeq 4000) was performed on formalin-fixed paraffin-embedded (FFPE) resected aortic tissue from 25 aortitis and 25 non-inflammatory aneurysm control patients undergoing surgical aortic repair (Table 1, Fig. 1). Differentially expressed genes (DEGs) between the two groups were identified while controlling for the potentially confounding effects (i.e., use of Statin, use of angiotensin converting enzyme inhibitors or angiotensin receptor blockers, and history of other rheumatic diseases). Protein-protein interactions from STRING and drug-gene pharmacological interaction information from DBIdb were used to construct DEG interactome and pharmacogenomic networks, respectively. In addition, DAVID was used to identify enriched biomolecular functions (Gene Ontology) for the gene sets from the DEGs, the DEG interactome network, and the pharmacogenomic network.
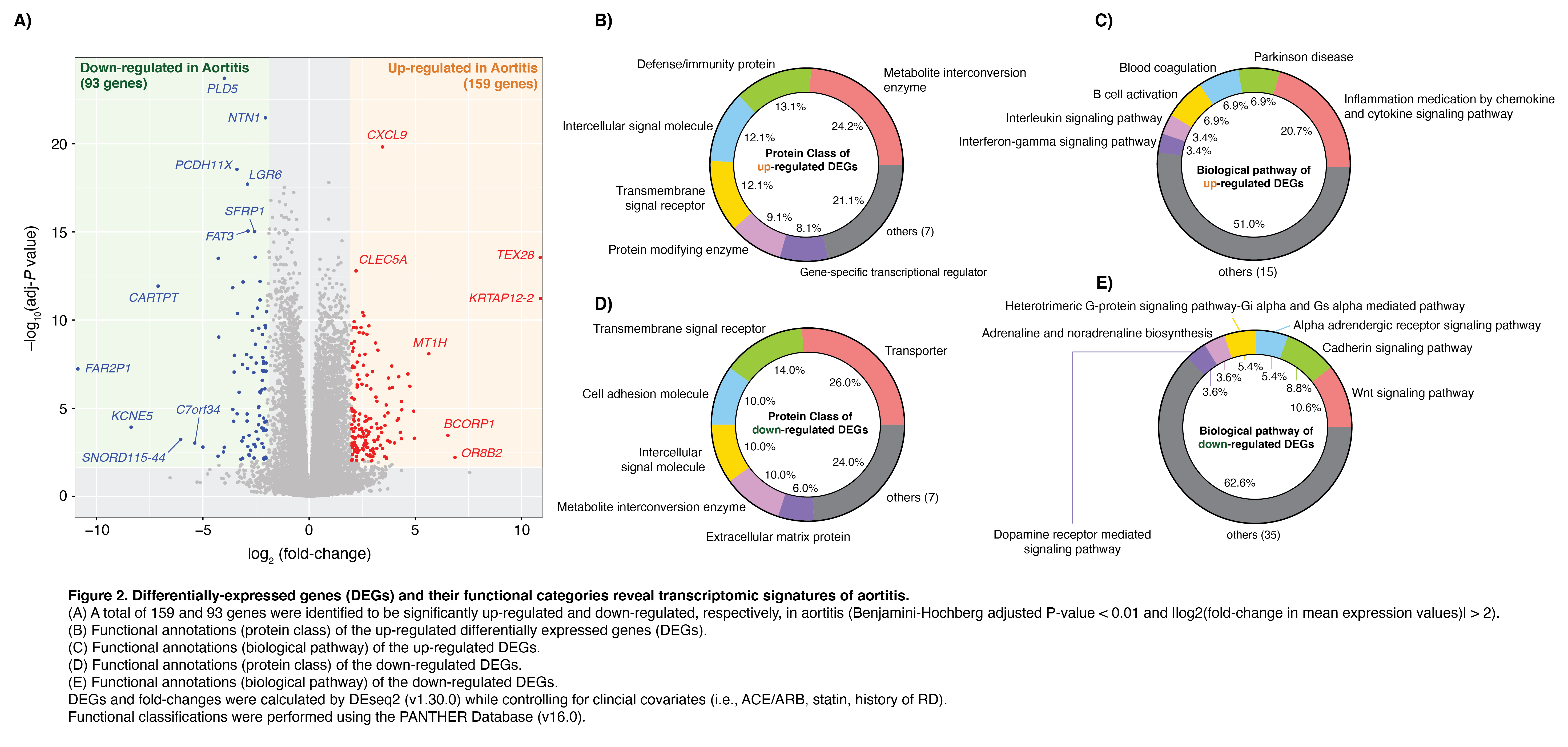
Results: We identified 159 and 93 genes to be significantly up-regulated and down-regulated, respectively, in aortitis (adj-P < 0.01, |log2(FC)| > 2) compared to non-inflammatory aortic tissue (Fig. 2). Up-regulated genes were enriched with functions related to immune response and inflammation, defense against bacteria, and interferon response pathways; while the down-regulated genes were enriched with plasma membrane and neuronal processes. Of note, the gene expression profiles of aortic tissue from patients with aortitis related to GCA (n=8) was no different than those of aortic tissue from patients with CIA (n=17). In aortitis, 30 out of 36 genes from the hub of the DEG interactome network were enriched with immune-related functions. Finally, our pharmacogenomic network analysis identified 16 genes that could be targeted with FDA-approved drugs.
Conclusion: Global transcriptome profiling on FFPE aortic specimens from patients with aortitis identified upregulated genes related to immune response and inflammation. Interestingly, we demonstrate for the first time that the transcriptomic signature of CIA is similar to that of GCA, suggesting that CIA may be a limited presentation of GCA. This study also facilitates the characterization of dysregulated molecular pathways in aortitis, and could provide a promising direction for biomarker discovery and treatment.
To cite this abstract in AMA style:
Hur B, Koster M, Jang J, Warrington K, Sung J. Global Transcriptomic Profiling Identifies Differential Gene Expression Signatures Between Inflammatory and Non-inflammatory Aortic Aneurysms [abstract]. Arthritis Rheumatol. 2021; 73 (suppl 9). https://acrabstracts.org/abstract/global-transcriptomic-profiling-identifies-differential-gene-expression-signatures-between-inflammatory-and-non-inflammatory-aortic-aneurysms/. Accessed .« Back to ACR Convergence 2021
ACR Meeting Abstracts - https://acrabstracts.org/abstract/global-transcriptomic-profiling-identifies-differential-gene-expression-signatures-between-inflammatory-and-non-inflammatory-aortic-aneurysms/