Session Information
Session Type: Abstract Session
Session Time: 1:00PM-2:30PM
Background/Purpose: The association between RA and the MHC is largely explained by five amino acid (AA) positions: DRB1 positions 11, 13, 71, and 74, HLA-B position 9, and DPB1 position 9. However, these prior studies are from populations genetically similar to individuals of European (EUR) ancestry, lacking data from individuals of African (AFR) ancestry. The Veteran Affairs (VA) Million Veteran Program (MVP), a diverse mega-biobank, provided an opportunity to fill this gap and compare RA genetics between individuals from AFR and EUR population.
Methods: VA MVP is a cohort of U.S. veterans ( >29% non-EUR ancestry) with linked electronic health record (EHR) and genotype data. Superpopulation membership (AFR, EUR) was based on genetic similarity to the 1000 Genomes Project reference panel. SNP2HLA was used to impute AAs and HLA alleles for seven genes. RA was defined with a phenotyping algorithm with a positive predictive value of 0.94 in both AFR and EUR. Omnibus testing across all AA positions controlling for age, sex, and population stratification estimated the total genetic effect on RA from polymorphic residue variation at an AA position. In a conditional haplotype manner, further rounds of omnibus testing were completed, conditioning on the most significant position of the prior round, to isolate a sequence of AA positions with independent effects. This was performed until no positions remained after Bonferroni correction (P< 0.05/1823). AFR-specific RA risk haplotypes were constructed from identified AA positions and effect sizes were estimated using multivariate logistic regression. HLA haplotype effect sizes were replicated using pooled data from two independent AFR RA cohorts (CLEAR: 348 RA, 1004 control; GENRA: 166 RA, 867 control).
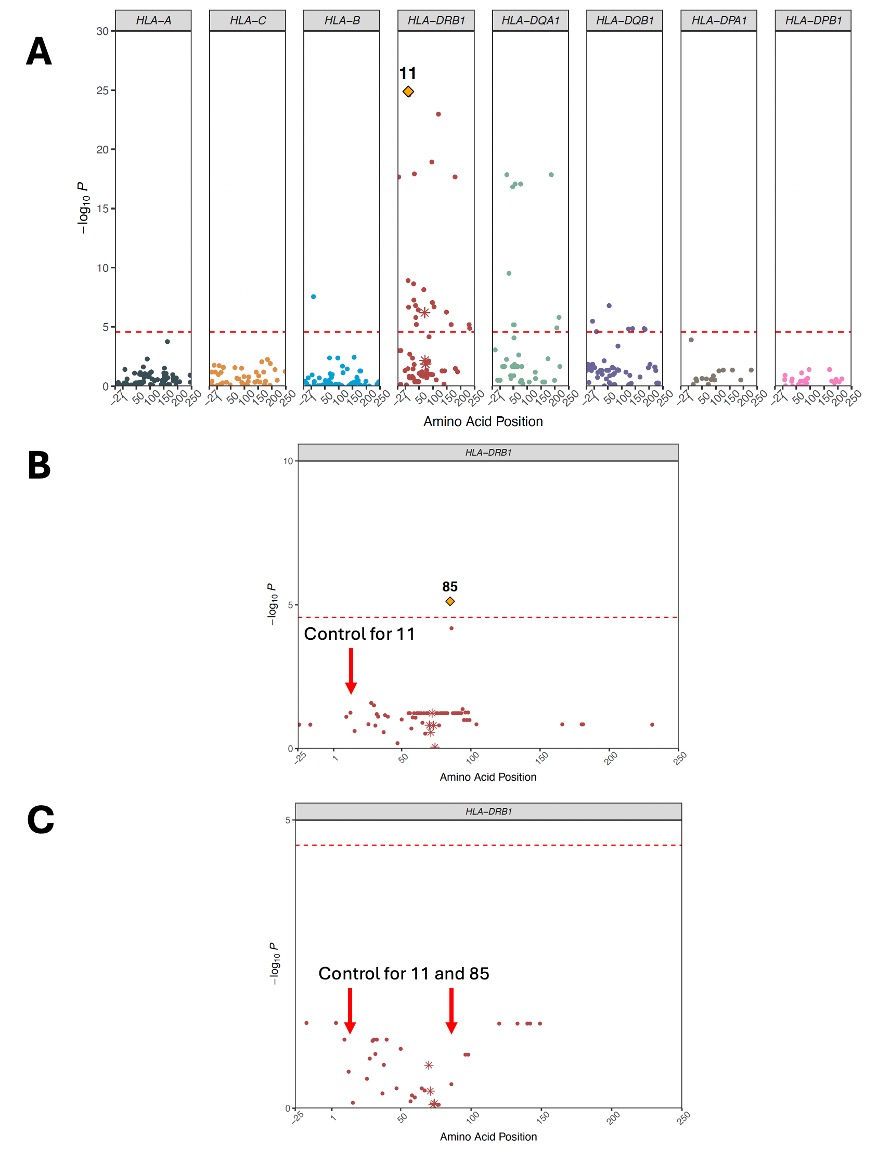
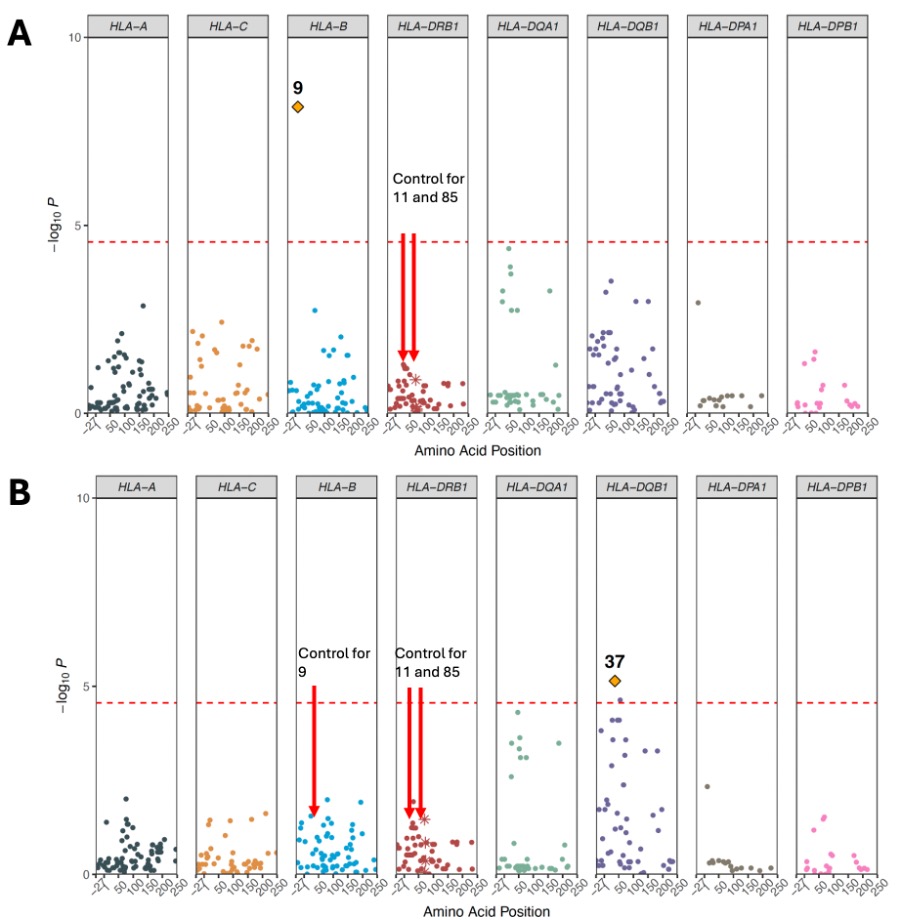
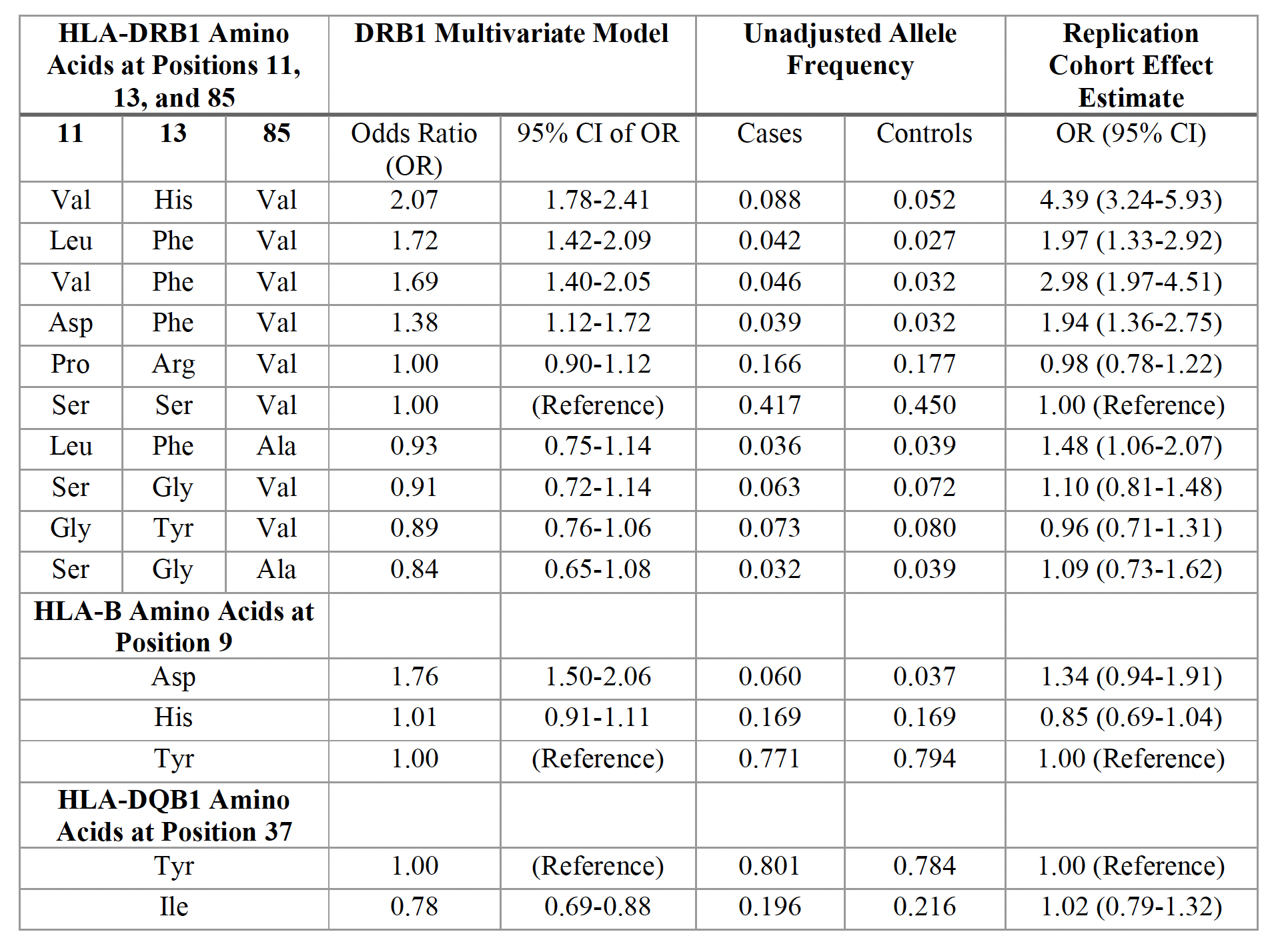
Results: We studied 1,522 RA and 116,568 non-RA Veterans from AFR with mean age 57 years, 14% female, and 70% seropositive. GWAS of the non-HLA region did not reveal novel associations. In line with prior studies, DRB1 positions 11 and 13 had the strongest association with RA (Figure 1A). In DRB1, controlling for position 11 identified position 85, distinct from secondary position 71 identified in EUR (Figure 1B). We then isolated the genetic effects of HLA-B position 9 and DQB1 position 37 on RA phenotype independent of DRB1 (Figure 2). Effect sizes of haplotypes from discovered positions across the MHC are reported in Table 1. The DRB1 haplotype with valine, histidine, and valine at positions 11, 13, and 85 respectively had the strongest association magnitude (OR=2.07, 95% CI=1.78-2.41). Effect sizes in the validation cohort were concordant with MVP (Table 1). The addition of previously unreported AFR haplotypes in this study to a genetic risk model using EUR-based haplotypes did not significantly improve model fit in EUR (Omnibus P=0.02) but significantly improved model fit in AFR (Omnibus P=3.07 x 10-9), suggesting distinct effects in AFR.
Conclusion: We identified distinct risk haplotypes that better characterize RA genetic architecture in AFR. Notably, some of the most significant haplotypes did not include the conventional ‘shared epitope’ alleles. These data contribute to constructing population-specific polygenic risk scores.
To cite this abstract in AMA style:
Zhang H, Sakaue S, Posner D, Cui J, Yang D, Budu-Aggrey A, Ho Y, Costa L, Matty R, Huang S, Monach P, Ishaigaki K, Maripuri M, Melley C, Tanukonda V, Sangar R, McDermott G, Jeffway M, Laufer V, Okada Y, Scott I, Bridges S, Cho K, Hong C, Huffman J, Cai T, Raychaudhuri S, Liao K. Genome-wide Association of Rheumatoid Arthritis in the African Ancestry Identifies HLA Amino Acid Polymorphisms of Risk [abstract]. Arthritis Rheumatol. 2024; 76 (suppl 9). https://acrabstracts.org/abstract/genome-wide-association-of-rheumatoid-arthritis-in-the-african-ancestry-identifies-hla-amino-acid-polymorphisms-of-risk/. Accessed .« Back to ACR Convergence 2024
ACR Meeting Abstracts - https://acrabstracts.org/abstract/genome-wide-association-of-rheumatoid-arthritis-in-the-african-ancestry-identifies-hla-amino-acid-polymorphisms-of-risk/