Session Information
Session Type: Poster Session C
Session Time: 10:30AM-12:30PM
Background/Purpose: Lupus nephritis (LN) is one of the most common and severe manifestations of systemic lupus erythematosus (SLE). We performed genome wide association studies (GWAS) for lupus nephritis and kidney function measures over time. We hypothesized that analyzing a person’s eGFR variability over time would be a good proxy for LN and improve power for detecting genetic loci for LN. We also used local ancestry estimation to facilitate inclusion of admixed individuals and generate ancestry specific estimates for LN outcomes.
Methods: We included SLE patients from several child and adult dedicated lupus databases and the Systemic Lupus International Collaborating Clinics (SLICC) cohort. All met American College of Rheumatology and/or SLICC SLE classification criteria and were genotyped on a multi-ethnic Illumina array. Ungenotyped SNPs were imputed to the Trans-Omics for Precision Medicine program (TopMed), and local ancestry of chromosomal information was estimated using RFMix and Tractor software. LN was defined by SLE criteria, with a subset confirmed by kidney biopsy. Kidney function (estimated glomerular filtration rate, eGFR) was calculated using the Schwartz Bedside formula for measures < 18 years and CKD-EPI for >18 years of age. Wilcoxon rank sum or Chi-square tests were used to compare characteristics such as kidney function between LN and Non-LN patients. We completed separate GWAS for the outcomes of LN, mean eGFR and eGFR variability over time (log of the mean absolute deviation from mean eGFR per participant), in marginal and multivariable adjusted regression models with sex, site and local principal components using Regenie. Local ancestry analysis was restricted to individuals of European, African and East Asian ancestry using Tractor. We meta-analyzed ancestry-specific results with METAL software (significance p< 5x10-8).
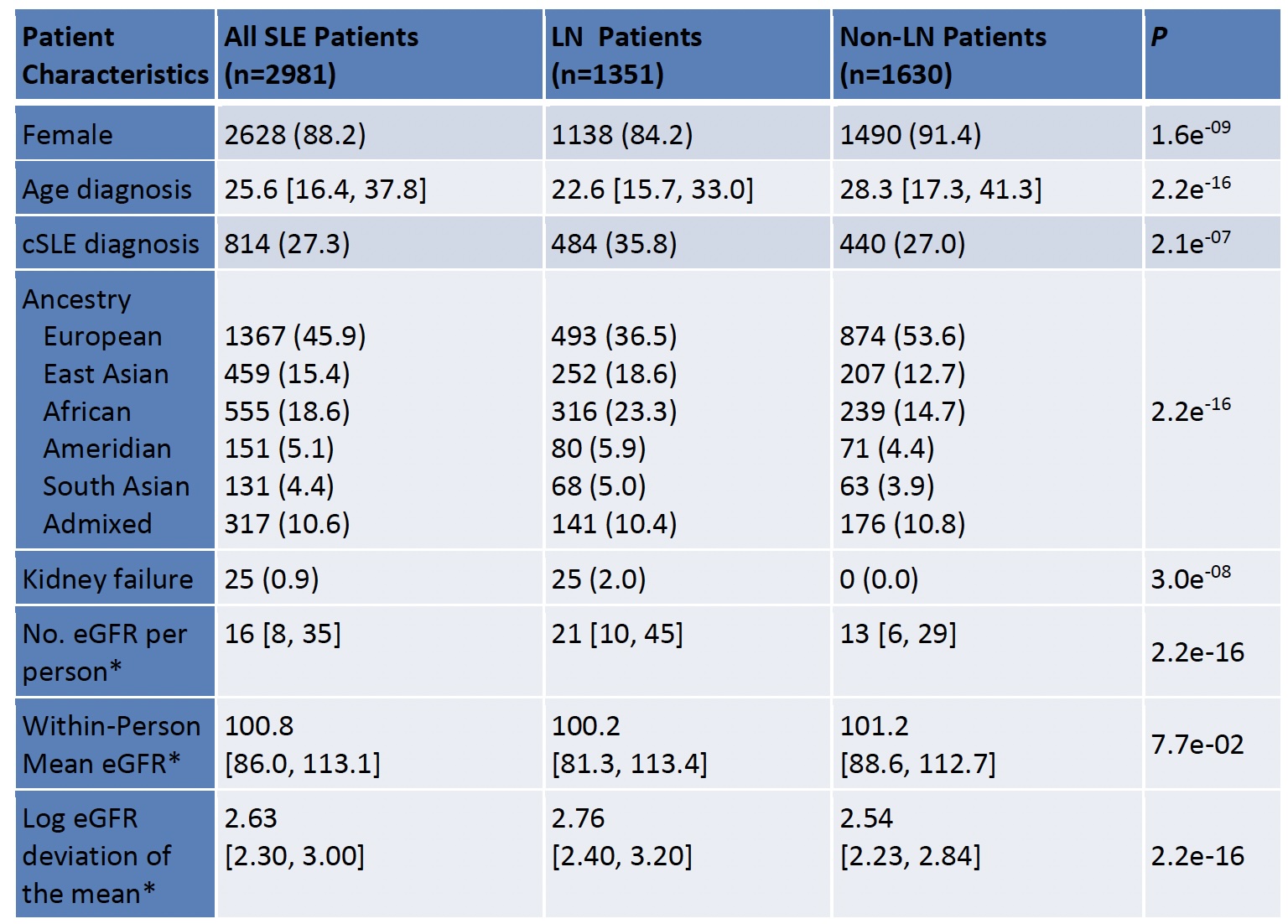
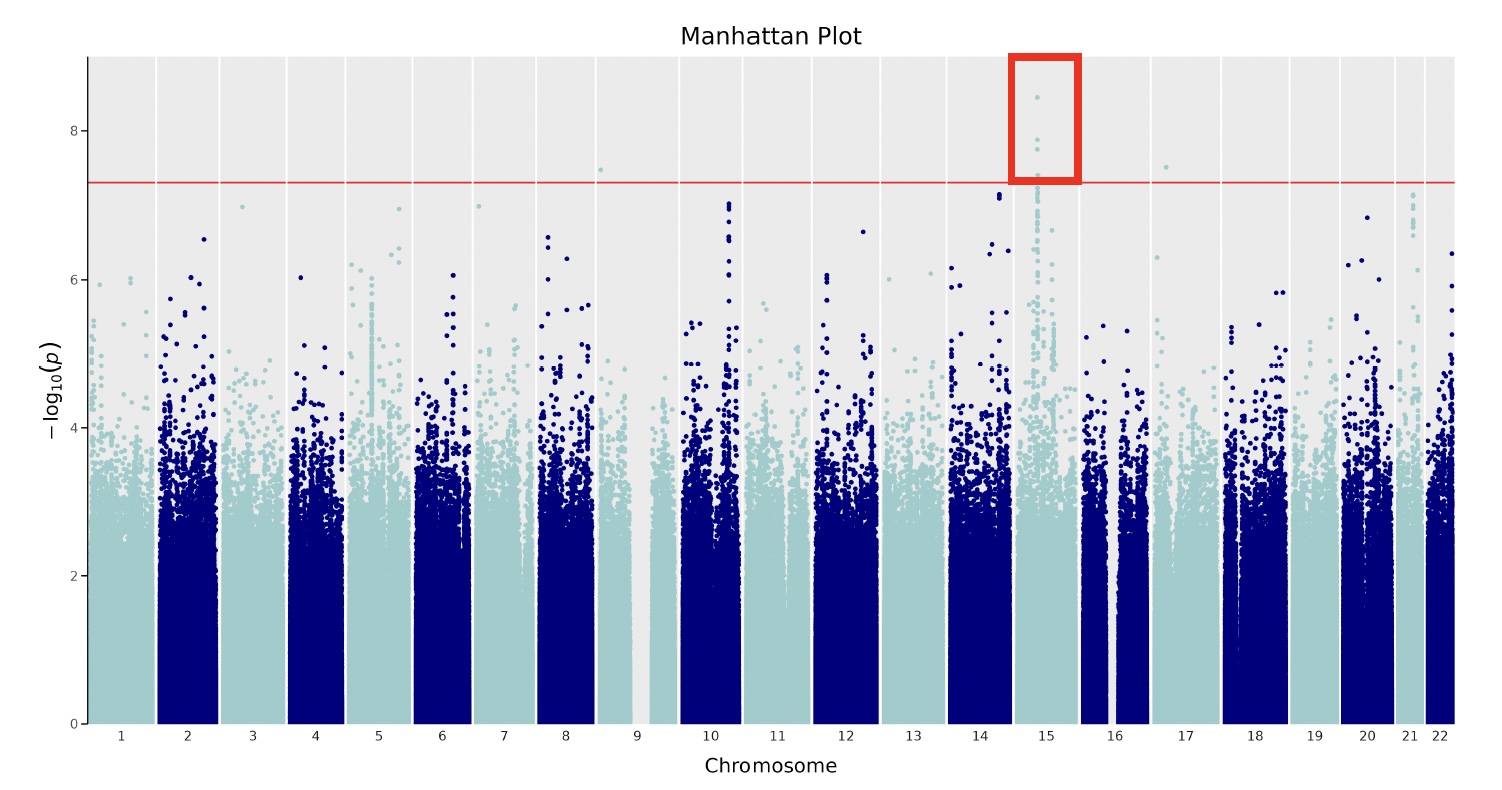
Results: We studied 2981 individuals with SLE, 88% female, 46% of European ancestry, 27% childhood-onset SLE, and 45% with LN (Table). Kidney failure was observed in 25 patients over a median follow-up time of 8.9 years (IQR: 4.1,14.8). Within-person eGFR was similar between people with and without LN, but eGFR variability was significantly greater in people with LN (P-value= 2.2e-16). Variability was calculated using a median of 16 [IQR: 8,35] eGFR measurements per person. GWAS of LN did not identify a significant LN locus, yet GWAS of eGFR variability demonstrated a significant peak on chromosome 15, downstream of SCH4 and intronic to SECISBP2L (Figure). The variant was found in a genomic region of African ancestry.
Conclusion: We completed GWAS of LN, eGFR mean and variability over time, generating ancestry specific estimates and identified a genome-wide significant locus for variability in measures of renal function over time, in a multiethnic cohort of children and adults with SLE. This locus was only found in an African ancestry portion of the genome. Variability in measures of renal function is correlated with LN, yet GWAS of LN did not identify significant loci. Future work includes repeating analyses to include all global ancestries, and to investigate the biologic link between the loci and LN.
To cite this abstract in AMA style:
Riedl Khursigara M, Gold N, Tang T, Cao J, Dominguez D, Klein-Gitelman M, Gladman D, Goldman D, Harvey E, Ishimori M, Jefferies C, Kamen D, Kamphuis S, Knight A, Lee C, Levy D, Noone D, Onel K, Peschken C, Petri M, Pope J, Pullenayegum E, Silverman E, Touma Z, Urowitz M, Wallace D, Wither J, Hiraki L. Genetics of eGFR Variability as a Proxy for Lupus Nephritis in Patients with Systemic Lupus Erythematosus [abstract]. Arthritis Rheumatol. 2024; 76 (suppl 9). https://acrabstracts.org/abstract/genetics-of-egfr-variability-as-a-proxy-for-lupus-nephritis-in-patients-with-systemic-lupus-erythematosus/. Accessed .« Back to ACR Convergence 2024
ACR Meeting Abstracts - https://acrabstracts.org/abstract/genetics-of-egfr-variability-as-a-proxy-for-lupus-nephritis-in-patients-with-systemic-lupus-erythematosus/