Session Information
Date: Sunday, November 10, 2019
Title: Pediatric Rheumatology – ePoster I: Basic Science, Biomarkers, & Sclerodermic Fever
Session Type: Poster Session (Sunday)
Session Time: 9:00AM-11:00AM
Background/Purpose: Localized scleroderma (LS) is a progressive autoimmune disease of the skin and underlying tissue that is characterized by an initial inflammatory infiltration which is followed by fibrosis and collagen deposition. When left unchecked LS causes significant functional disability and disfigurement, especially in developing children. Histopathologic review of LS skin biopsies typically shows a stronger inflammatory or fibrotic pattern depending on disease stage. Our goal was to determine the transcriptome within inflammatory vs. fibrotic tissue to identify potential molecular targets using RNA sequencing (RNAseq) compared to histology scoring of inflammatory infiltrates vs. collagen deposition/fibrosis. Differentially expressed genes (DEGs) in LS patients were compared to skin histopathological features to determine correlation.
Methods: RNAseq was performed on paraffin-embedded skin (n=15 LS, n=2 pediatric healthy) using the Illumina HTS and TrueSeq Access library preparation. Paired-end RNAseq data was aligned using STAR and analyzed for DEGs using DESeq2. Genes were analyzed using DEG cutoffs of log fold change > ±2.0, adjusted p< 0.05, and a false discovery rate (FDR) cutoff of < 0.05 for Ingenuity Pathway Analysis (IPA) and enrichment software (GSEA©). Skin biopsies were reviewed by 2 blinded pathologists who determined 3 areas of inflammatory infiltrate per skin layer (papillary dermis, upper and lower reticular dermis) and counted total inflammatory cells (lymphocytes and plasma cells) per infiltrate, which determined a categorical inflammation score. Similar scoring was also developed for fibrosis, where 3 areas were chosen per skin layer and 3 measurements of collagen bundle thickness were taken. The average of these measurements was used for fibrotic scoring. Spearman’s correlation between genes of interest and histology scoring was performed using GraphPad Prism. Results: The degree of inflammatory cell infiltrate significantly correlated with both inflammatory (IFNγ, immunoglobulin, and T-cell activation, rs > ±0.5, p< 0.01) and fibrotic (IL-12 mediated signaling, and DNA damage response) (rs > ±0.5, p< 0.05) genes (Table 1). Collagen thickness did not correlate with typical fibrotic genes, but did correlate (rs > ±0.4, p< 0.05) with upregulated inflammatory genes, such as those associated with interferon signaling and T-cell activation (Table 1). Further support of IFN-related signaling, T lymphocyte activation, and cytokine production was demonstrated with IPA. Conclusion: The identified immune pathways and genes corresponding with the LS skin inflammatory infiltrate indicate a unique genetic signature present during active disease with moderate-severe lymphoplasmacytic infiltrate and collagen thickness. This may indicate that fibrosis occurring in later stages of LS is an inflammation-driven process. Further investigation into the relationship and functions of these genes is underway and will aid in advancing treatment options for LS patients.
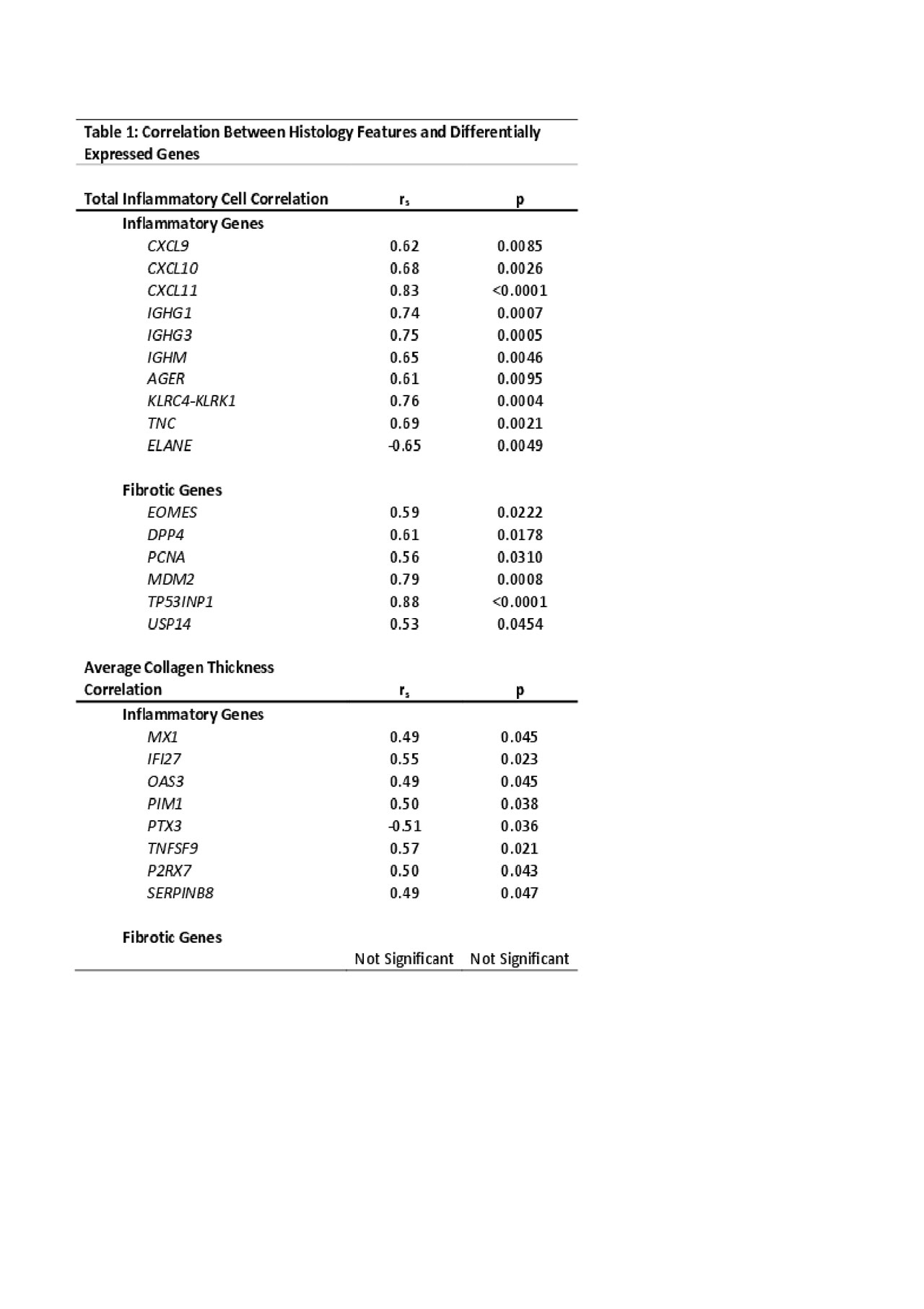
Table 1 Correlation Analysis Between Histology Features and Differentially Expressed Genes FINAL
To cite this abstract in AMA style:
Schutt C, Mirizio E, Salgado C, Reyes-Mugica M, Schollaert K, Torok K. Genetic Signatures Support Inflammation Driven Fibrosis in Localized Scleroderma [abstract]. Arthritis Rheumatol. 2019; 71 (suppl 10). https://acrabstracts.org/abstract/genetic-signatures-support-inflammation-driven-fibrosis-in-localized-scleroderma/. Accessed .« Back to 2019 ACR/ARP Annual Meeting
ACR Meeting Abstracts - https://acrabstracts.org/abstract/genetic-signatures-support-inflammation-driven-fibrosis-in-localized-scleroderma/
