Session Information
Session Type: Poster Session C
Session Time: 9:00AM-11:00AM
Background/Purpose: Juvenile idiopathic arthritis (JIA) encompasses multiple forms of pediatric autoimmune arthritis. Research studies in JIA are complicated by disease heterogeneity and difficulties gathering large numbers of patients for longitudinal studies. Electronic health records (EHRs) store nearly all clinical data in one central location, are used widely across institutions, and are becoming a major source of data for studying diseases in a real-world population with longitudinal data. In this study, we use an algorithm to accurately identify a JIA cohort from an EHR linked to a DNA biobank and perform genetic studies. The goal of this work is to establish a cost-effective method for performing JIA studies in a cohort with longitudinal clinical data.
Methods: A portable algorithm was used to identify JIA patients using a combination of ICD-9-CM and ICD-10-CM codes, age criteria, and exclusion of ICD codes associated with connective tissue disease. Classification of JIA status by the algorithm was compared to true cases as determined by a rheumatology clinic note documenting JIA before 20 years old. Individuals with ICD-9-CM and ICD-10-CM codes for well child visits and without JIA-associated ICD codes or use of disease-modifying and biologic medications used to treat JIA were classified as controls. Identified cases and controls with existing genotype data from a Multi-Ethnic Genotyping Array (MEGA-ex) were included. A genome-wide association study (GWAS), adjusted for 10 principal components, was performed. S-PrediXcan was used to assess associations between JIA and genetically predicted gene expression in placenta and 48 GTEx (v7) tissues. Functional Mapping and Annotation (FUMA) of GWAS was used for annotation of single-nucleotide polymorphisms (SNPs) and to test for enrichment of predefined biological pathways. Significance was determined by a Bonferroni corrected p-value.
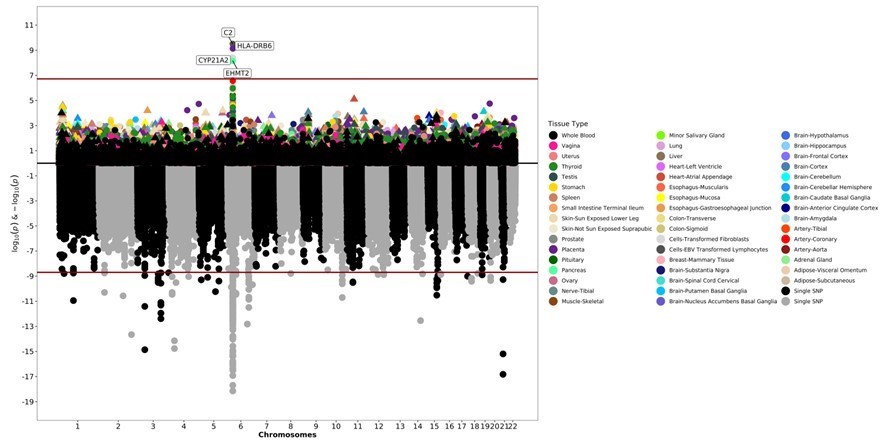
Results: The JIA algorithm identified 1,379 cases with a positive predictive value of 0.98 from a cohort of over 3.5 million records. There were 229 cases and 2,798 controls with genotype data. Cases were 68% female and 84% white while controls were 58% female and 48% white. GWAS identified numerous significant SNPs, the majority of which were on chromosome 6. Significant results of note include SNPs previously associated with JIA (rs2395148, rs7775055) (Figure 1). FUMA analysis identified 29 genomic risk loci and 33 lead SNPs. Canonical pathways, GO biological processes, and GO molecular functions were enriched for important processes in the immune system. S-PrediXcan identified 4 genes (C2, HLA-DRB6, CYP21A2, and EHMT2) predicted to be differentially expressed in JIA (Figure 1).
Conclusion: A portable algorithm can identify JIA patients in a real-world cohort with longitudinal clinical data. This phenotype is highly accurate and leads to biologically plausible genetic associations with JIA. This is a cost-effective method for studying JIA and its genetic associations. This approach can be expanded to generate larger cohorts across multiple institutions.
The bottom of the graphic is a Manhattan plot which displays significant SNPs from GWAS. The top of the graphic is the results from S-PrediXcan, with symbols now representing entire genes and their genetically determined expression levels. The x-axis are chromosomes. The y-axis is log and negative log p-values from the GWAS and S-PrediXcan analyses. Colors correspond to specific tissues.
To cite this abstract in AMA style:
Jasper E, Gangireddy S, Ong H, Hellwege J, Edwards T, Velez Edwards D, Wei W, Patrick A. Genetic Associations in Juvenile Idiopathic Arthritis Determined with an Electronic Health Record Based Approach [abstract]. Arthritis Rheumatol. 2023; 75 (suppl 9). https://acrabstracts.org/abstract/genetic-associations-in-juvenile-idiopathic-arthritis-determined-with-an-electronic-health-record-based-approach/. Accessed .« Back to ACR Convergence 2023
ACR Meeting Abstracts - https://acrabstracts.org/abstract/genetic-associations-in-juvenile-idiopathic-arthritis-determined-with-an-electronic-health-record-based-approach/