Session Information
Session Type: Abstract Submissions (ACR)
Background/Purpose: Systemic Lupus Erythematosus (SLE) is an autoimmune disorder predominately affecting females in the reproductive age range. Estrogen is present at higher levels in this population compared to pre-pubertal and postmenopausal women and has been shown to influence immune cell function by regulating the expression of multiple genes through activation of estrogen receptors (ER) a and b. Recent work using a chromatin immunoprecipitation with ERa has shown that this list may be much larger than what has been characterized to date. In this study, the effects of E2 were examined by comparing gene array expression in peripheral blood mononuclear cells (PBMCs) from premenopausal women with SLE and in healthy premenopausal individuals.
Methods: SLE patients meeting the revised criteria of the American College of Rheumatology and healthy volunteers were recruited through approved IRB protocols. Whole blood was collected into heparnized tubes and PBMCs were isolated. Cells were cultured with or without 10 nM of 17b-estradiol (E2) for 48 hours. PBMCs were then collected and purified as total RNA and submitted for gene array analysis using HG-U133 Affymetrix® Human Gene Chips. Untreated PBMC arrays served as the internal baseline control for each individual sample and was subtracted from the E2-treated expression values; thus, only the estrogen mediated effect was reported. Data was analyzed using the Multiplot function within the Genepattern software program and with Ingenuity Pathway Analysis Software.
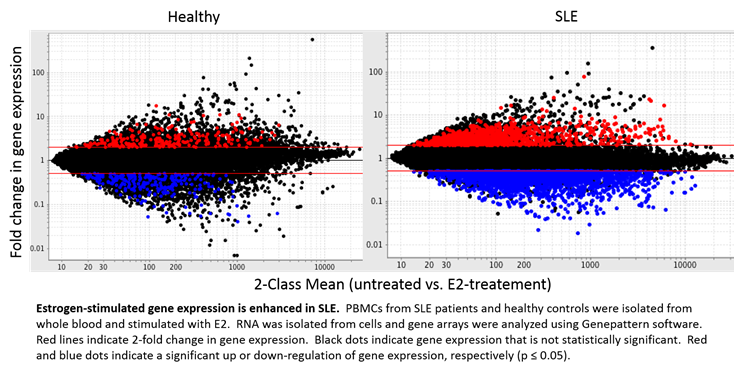
Results: While over 1000 genes were significantly up-regulated over 2-fold in SLE samples when treated with E2, only 236 were identified in corresponding healthy controls. Furthermore, E2 stimulation significantly down-regulated 2530 genes in PBMCs from SLE patients, but only 244 in healthy samples under the same conditions. In concordance, 525 and 1629 genes were found to be uniquely up or down-regulated with E2 treatment in SLE, respectively. Significant E2-mediated regulation of several methyltransferases, including PCMTD1 and RG9MTD3, was identified in this analysis. Using Ingenuity Pathway Analysis Software, our results identified these gene sets to be associated with several pathways, including post-translational modification.
Conclusion: These results establish a clear estrogen effect over many genes to produce an estrogen signature, including several genes known to be associated with post-translational modification, and further reveals enhanced regulation in SLE patients when compared to healthy controls.
Disclosure:
S. Amici,
None;
N. A. Young,
None;
L. C. Wu,
None;
M. Guerau,
None;
W. N. Jarjour,
None.
« Back to 2014 ACR/ARHP Annual Meeting
ACR Meeting Abstracts - https://acrabstracts.org/abstract/gene-array-analysis-reveals-unique-estrogen-signature-in-peripheral-blood-mononuclear-cells-of-patients-with-systemic-lupus-erythematosus/