Session Information
Date: Tuesday, October 28, 2025
Title: Plenary III (1722–1727)
Session Type: Plenary Session
Session Time: 8:45AM-9:00AM
Background/Purpose: Systemic sclerosis (SSc) is characterized by vasculopathy, progressive fibrosis of skin and internal organs, and autoimmunity. Notably, African American (AA) patients with SSc exhibit a more severe vascular phenotype and poorer outcomes than European Americans. We hypothesized that common variants in SSc tag gene regions enriched for rare, functional variants cause increased disease burden in AAs with SSc.
Methods: Exome and targeted sequencing were performed in discovery and replication cohorts comprising 969 AA SSc patients and 771 AA controls, focusing on 32 SSc-associated genes. Two NOTCH4 variants were examined in lymphoblastoid cell lines (LCL) using RT-PCR, ELISA, and flow cytometry. SSc skin single-cell RNA-Seq and publicly available bulk gene expression datasets were analyzed. Endothelial cell (EC) lines and variant-carrying ECs from SSc patients (pECs) were used to assess angiogenesis using tube formation assays and to study endothelial-to-mesenchymal transition (EndoMT) following NOTCH4 stimulation or inhibition. A Tie2-tTA;TRE-Notch4* mouse model with inducible, EC-specific expression of active Notch4 was used to study EndoMT.
Results: Gene-based testing identified NOTCH4 association with SSc (P=1.6×10-7) particularly in AAs with severe vasculopathy (P=3.5×10-7). The risk haplotype “TA”, defined by the missense (c.2824C >T) and promoter (c.-117G >A) variants, was enriched in AAs with SSc (11%) and increased the risk of having severe Raynaud’s, scleroderma renal crisis (SRC), and pulmonary arterial hypertension (PAH) (OR=10.6, 95% CI 2-56) (Fig. 1A-B). The population attributable risk due to this haplotype in AAs with SSc was 2.6%, 52-fold higher than in European Americans (Fig. 1C). The c.-117A allele bound glucocorticoid receptor and increased NOTCH4 protein expression (Fig. 1D-E). SSc skin samples with c.-117G >A variant had a higher NOTCH4 expression than those with the G allele (Fig. 1F). The c.2824T allele upregulated NOTCH4 signaling in LCLs (Fig. 1G-H), confirmed in vivo by elevated HES1 transcripts in c.2824T vs. c.2824C SSc samples (Fig. 1I). scRNA-Seq data for SSc skin revealed that NOTCH4 is predominantly expressed by ECs (Fig. 2 A-B) especially arterial, mature capillary and tip cells (Fig 2 C-D). NOTCH4 stimulation in the EC line and c.-117A allele-carrying pECs led to decreased tube formation (an angiogenesis indicator) (Fig. 2E-H) and increased EndoMT (Fig. 2K-L). NOTCH4 overexpression in Tie2-tTA;TRE-Notch4* mutant mice also increased Acta2 (EndoMT marker) expression in ECs (Fig. 2I-J). Nailfold capillary abnormalities, decreased angiogenesis, and fibrosis of the vascular lumen are commonly seen in SSc. Inhibition of NOTCH4 using genetic knockdown, blocking antibody, or the FDA-approved drug Nirogacestat restored angiogenesis (Fig. 2 E-H) and normalized EndoMT in ECs (Fig. 2 K-L).
Conclusion: Functional variants in NOTCH4 gene are associated with SSc pathogenesis and vasculopathy and may explain the higher disease burden of SSc in AAs. Blocking NOTCH4 signaling restored angiogenesis and EndoMT, highlighting its potential as a therapeutic target in restoring angiogenesis in SSc and connective tissue diseases.
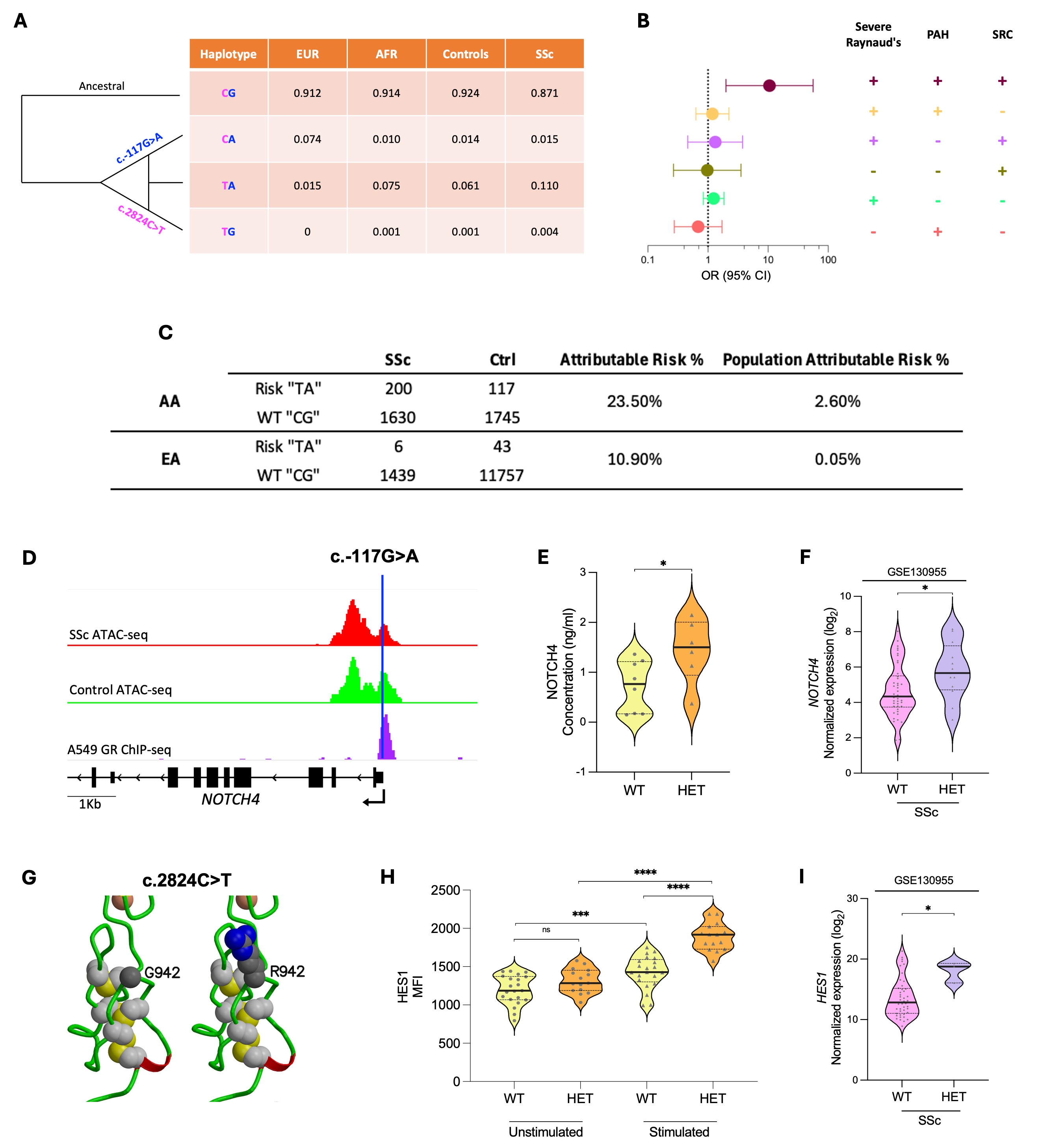 Fig. 1. A. Frequencies of c.2824C>T and c.-117G>A haplotypes in SSc cases, controls and reference European (EUR) (n=136) and African (AFR) (n=471) ancestral populations. B. Odds ratios for the risk “TA” haplotype vs. wildtype “CG” across SSc vascular phenotypes. C. Population Attributable risk for the NOTCH4 haplotype in African Americans (AA) and European Americans (EA). D-F show c.-117G>A data. G-I show c.2824C>T data. D. ATAC-seq signal at the NOTCH4 locus in primary endothelial cells from SSc patients and healthy controls (GSE163199) and the GR-a ChIP-seq in A549 cells with c.-117G>A marked in blue. E. NOTCH4 ELISA data in unstimulated LCLs wild type (WT) or Heterozygous (HET) for c.-117G>A. F. NOTCH4 expression in SSc skin, stratified by the c.-117G>A variant (GSE130955). G. c.2824C>T alters glycine to arginine at position 942. H. Flow cytometry data for HES1 before and after stimulation of NOTCH4 with DLL4 ligand, in LCLs WT or HET for the c.2824C>T variant. I. HES1 expression in SSc skin, stratified by the c.2824C>T variant (GSE130955).
Fig. 1. A. Frequencies of c.2824C>T and c.-117G>A haplotypes in SSc cases, controls and reference European (EUR) (n=136) and African (AFR) (n=471) ancestral populations. B. Odds ratios for the risk “TA” haplotype vs. wildtype “CG” across SSc vascular phenotypes. C. Population Attributable risk for the NOTCH4 haplotype in African Americans (AA) and European Americans (EA). D-F show c.-117G>A data. G-I show c.2824C>T data. D. ATAC-seq signal at the NOTCH4 locus in primary endothelial cells from SSc patients and healthy controls (GSE163199) and the GR-a ChIP-seq in A549 cells with c.-117G>A marked in blue. E. NOTCH4 ELISA data in unstimulated LCLs wild type (WT) or Heterozygous (HET) for c.-117G>A. F. NOTCH4 expression in SSc skin, stratified by the c.-117G>A variant (GSE130955). G. c.2824C>T alters glycine to arginine at position 942. H. Flow cytometry data for HES1 before and after stimulation of NOTCH4 with DLL4 ligand, in LCLs WT or HET for the c.2824C>T variant. I. HES1 expression in SSc skin, stratified by the c.2824C>T variant (GSE130955).
.jpg) Fig. 2. A. UMAP plot of subclustered skin cells of SSc and control subjects with colors denoting different subpopulation. B. Density plot for NOTCH4 expression, primarily in endothelial cells, with lighter color indicating higher expression. C. UMAP plot of reclustered endothelial cells in SSc and control subject. D. Density plot for NOTCH4 expression, primarily in arterial, mature capillary and tip endothelial cell cluster, with lighter color indicating higher expression. E. Decreased tube formation upon NOTCH4 activation by its ligand DLL4 in endothelial cell line, unaffected by prior NOTCH4 knockdown. F. Restoration of normal tube formation after inhibition of NOTCH4 signaling using anti-NOTCH4 antibody, DAPT, or Nirogacestat prior to DLL4 stimulation. G-H. Rescued tube formation in SSc primary endothelial cells (WT and HET for c.-117G>A variant) after blocking NOTCH4 signaling with anti-NOTCH4 antibody or Nirogacestat. I. Co-staining of Acta2 (red), CD31 (green) and Erg (white) in thin-walled vessels of constitutively Notch4* expressing brain endothelial cells in Tie2-tTA;TRE-Notch4* mutant mice and Tie2-tTA littermate control mice. J. Quantification of thin-walled vessels co-stained for Acta2 (EndoMT marker) and CD31 in constitutively Notch4* expressing brain endothelial cells in Tie2-tTA;TRE-Notch4* mutant mice and Tie2-tTA littermate control mice. K. Increase in EndoMT markers NOTCH3 and SNAI2 following DLL4-induced NOTCH4 stimulation; restored to baseline levels with Nirogacestat. L. Decrease in ACTA2, COL1A1, and SNAI1 expression in primary endothelial cells from SSc patients (Het for c.-117G>A) after Nirogacestat treatment.
Fig. 2. A. UMAP plot of subclustered skin cells of SSc and control subjects with colors denoting different subpopulation. B. Density plot for NOTCH4 expression, primarily in endothelial cells, with lighter color indicating higher expression. C. UMAP plot of reclustered endothelial cells in SSc and control subject. D. Density plot for NOTCH4 expression, primarily in arterial, mature capillary and tip endothelial cell cluster, with lighter color indicating higher expression. E. Decreased tube formation upon NOTCH4 activation by its ligand DLL4 in endothelial cell line, unaffected by prior NOTCH4 knockdown. F. Restoration of normal tube formation after inhibition of NOTCH4 signaling using anti-NOTCH4 antibody, DAPT, or Nirogacestat prior to DLL4 stimulation. G-H. Rescued tube formation in SSc primary endothelial cells (WT and HET for c.-117G>A variant) after blocking NOTCH4 signaling with anti-NOTCH4 antibody or Nirogacestat. I. Co-staining of Acta2 (red), CD31 (green) and Erg (white) in thin-walled vessels of constitutively Notch4* expressing brain endothelial cells in Tie2-tTA;TRE-Notch4* mutant mice and Tie2-tTA littermate control mice. J. Quantification of thin-walled vessels co-stained for Acta2 (EndoMT marker) and CD31 in constitutively Notch4* expressing brain endothelial cells in Tie2-tTA;TRE-Notch4* mutant mice and Tie2-tTA littermate control mice. K. Increase in EndoMT markers NOTCH3 and SNAI2 following DLL4-induced NOTCH4 stimulation; restored to baseline levels with Nirogacestat. L. Decrease in ACTA2, COL1A1, and SNAI1 expression in primary endothelial cells from SSc patients (Het for c.-117G>A) after Nirogacestat treatment.
Disclosures: U. Kaundal: None; P. Tsou: None; M. Sahu: None; M. Huang: None; S. Boyden: None; C. Woodford: None; D. Shriner: None; S. Safran: None; Y. Zhou: None; X. Zhang: None; Y. Kunishita: None; A. Shah: None; M. Mayes: Argenx, 2, AstraZeneca, 5, atyr, 5, Boehringer-Ingelheim, 5, Bristol-Myers Squibb(BMS), 1, 5, h, 5, Novartis, 2, prometheus merck, 5; A. Doumatey: None; A. Bentley: None; R. Domsic: None; T. Medsger, Jr: None; P. Ramos: None; R. Silver: None; V. Steen: None; J. Varga: None; V. Hsu: None; L. Saketkoo: Abbvie, 6, Argenx, 1, 2, 5, aTyr Pharmaceuticals, 12,, 1, 5, Boehringer-Ingelheim, 2, 5, 6, CSL Behring, 5, EMD Serono, 2, 5, Horizon, 5, Johnson & Johnson, 6, Kadmon, 5, Kinevant, 12,, 2, 5, Mallinckrodt, 1, 2, 5, Novartis, 1, 2, 5, Priovant, 5; E. Schiopu: None; J. Gordon: Cumberland, 5, Prometheus/Merck, 5; L. Criswell: None; H. Gladue: None; C. Derk: None; E. Bernstein: AstraZeneca, 5, aTYR, 5, Boehringer-Ingelheim, 2, 5, Bristol-Myers Squibb(BMS), 5, Cabaletta Bio, 5, Synthekine, 2; S. Bridges: None; V. Shanmugam: None; L. Chung: AbbVie/Abbott, 1, Boehringer-Ingelheim, 1, CRISPR Therpeutics, 2, Cure Ventures, 2, jade, 2, Kyverna, 6, Mediar, 1, 2; S. Kafaja: None; M. TROJANOWSKI: None; B. Korman: None; J. Thomas: None; S. Dell'orso: None; d. Randazzo: None; A. Adeyemo: None; E. Remmers: None; P. Schwartzberg: None; I. Aksentijevich: In Vitro Diagnostics, 9; C. Rotimi: None; F. Wigley: None; R. Wang: None; F. Boin: Adicet Bio, 2, Boehringer Ingelheim, 1, Janssen Pharmaceuticals, 6; D. Khanna: Argenx, 2, AstraZeneca, 2, Boehringer-Ingelheim, 2, Bristol-Myers Squibb(BMS), 2, Cabaletta, 2, Novartis, 2, UCB, 2, Zura Bio, 2; R. Lafyatis: AbbVie/Abbott, 2, Advarra/GSK, 12, Independent Data Safety Monitoring Committees, Boehringer-Ingelheim, 2, Bristol-Myers Squibb(BMS), 2, 5, EMD Sereno, 2, Formation, 2, 5, Genentech, 12, Independent Data Safety Monitoring Committees, Genentech/Roche, 2, Mediar, 2, Merck/MSD, 2, Moderna, 5, Modumac Therapeutics Inc., 12, President and holds stock, Morphic, 2, Pfizer, 5, Regeneron, 5, Sanofi, 2, Third Rock Ventures, 2, Thirona Bio, 2, Zag Bio, 2; D. Kastner: In Vitro Diagnostics, 12, NIH Licensing Agreement; P. Gourh: None; E. Stenson: None; T. Talley: None; K. Gudapati: None; J. Wong: None; R. Jan: None; A. Goldberg: None; J. Mullikin: None.
To cite this abstract in AMA style:
Kaundal U, Tsou P, Sahu M, Huang M, Boyden S, Woodford C, Shriner D, Safran S, Zhou Y, Zhang X, Kunishita Y, Shah A, Mayes M, Doumatey A, Bentley A, Domsic R, Medsger, Jr T, Ramos P, Silver R, Steen V, Varga J, Hsu V, Saketkoo L, Schiopu E, Gordon J, Criswell L, Gladue H, Derk C, Bernstein E, Bridges S, Shanmugam V, Chung L, Kafaja S, TROJANOWSKI M, Korman B, Thomas J, Dell'orso S, Randazzo d, Adeyemo A, Remmers E, Schwartzberg P, Aksentijevich I, Rotimi C, Wigley F, Wang R, Boin F, Khanna D, Lafyatis R, Kastner D, Gourh P, Stenson E, Talley T, Gudapati K, Wang J, Jan R, Goldberg A, Mullikin J. Functional NOTCH4 Variants Drive Vasculopathy and Fibrosis in Systemic Sclerosis. [abstract]. Arthritis Rheumatol. 2025; 77 (suppl 9). https://acrabstracts.org/abstract/functional-notch4-variants-drive-vasculopathy-and-fibrosis-in-systemic-sclerosis/. Accessed .« Back to ACR Convergence 2025
ACR Meeting Abstracts - https://acrabstracts.org/abstract/functional-notch4-variants-drive-vasculopathy-and-fibrosis-in-systemic-sclerosis/
