Session Information
Date: Sunday, October 21, 2018
Title: T Cell Biology and Targets in Autoimmune and Inflammatory Disease Poster
Session Type: ACR Poster Session A
Session Time: 9:00AM-11:00AM
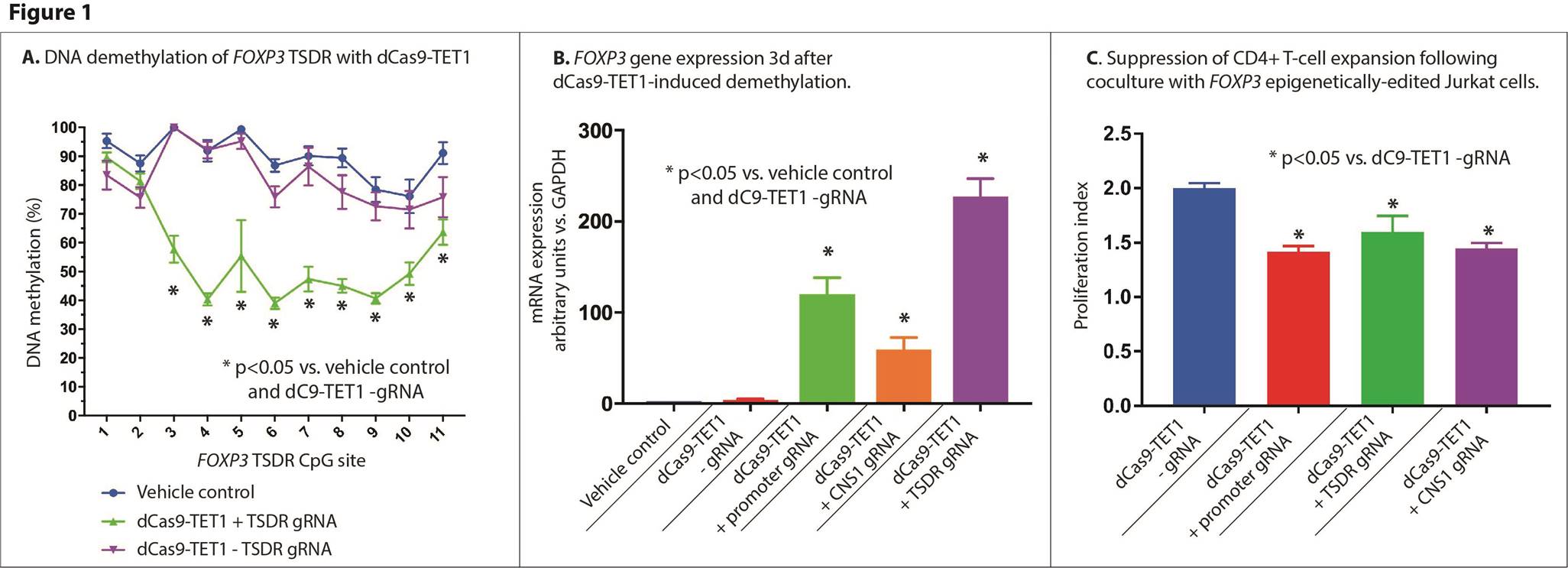
Background/Purpose: Defects in the number and function of naturally-occurring regulatory T cells (Tregs) have been identified in a variety of autoimmune diseases. The development of Tregs is marked by a variety of epigenetic modifications, most closely associated with demethylation of the FOXP3 gene. Transfection of T cells with FOXP3 transgenes has resulted in conversion to a Treg phenotype; however, the effects on cellular function of direct epigenetic editing of FOXP3 has not yet been evaluated.
Methods: Guide RNA (gRNA) sequences targeting the human FOXP3 promoter, TSDR, and CNS1 region were designed. An epigenetic editing SUNTAG (dCas9 + TET1) construct was used to demethylate genomic regions. Constructs containing dCas9 + TET1 + guide RNA sequences + fluorescent reporter were transfected by electroporation into Jurkat cells and cultured for 24 hours, then enriched by flow sorting. FOXP3 and CTLA4 gene expression was determined by qPCR and FOXP3-TSDR DNA methylation quantified by bisulfite pyrosequencing 3 days after transfection. Primary CD4+ T cells were isolated from human peripheral blood, stained with CellTrace reagent and combined with FOXP3 epigenetically-edited Jurkat cells in a 1:1 ratio and stimulated with anti-CD3/anti-CD28 beads and 30U/mL rhIL-2 for 5 days. Suppression of T cell division was determined by flow cytometry.
Results: Epigenetic editing constructs targeting the FOXP3 TSDR reduced DNA methylation (TSDR region, transfected: 55%±2 mean±SEM vs. vehicle: 91±3, p=8E-5) (Figure 1), whereas constructs targeting the promoter and CNS1 did not reduce TSDR methylation. All gRNAs increased FOXP3 expression (TSDR-targeted vehicle: 0.7±0.2, dC9-TET1+TSDR gRNA 230±10 relative units vs. GAPDH, p<0.0001) (promoter-targeted: vehicle: 0.7±0.2, dC9-TET1+Prom. gRNA 120±20, p=0.02), (CNS1-targeted vehicle: 0.7±0.2, dC9-TET1 +CNS2 gRNA 59±10, p=0.001) (Figure 1). Furthermore, epigenetic editing of FOXP3 resulted in increased expression of the Treg-related gene CTLA4 (FOXP3 prom.-targeted non-significant, dC9-TET1+FOXP3 TSDR gRNA 6.4±0.6 vs. vehicle: 1.6±0.4, p=0.003; dC9-TET1+FOXP3+CNS1 gRNA: 3.9±5 vs. vehicle: 0.8±0.3, p=0.009). Epigenetically edited cells resulted in suppression of naïve T-cell proliferation by 20-30% (dC9+TET1+prom. gRNA=29% avg. suppression, p=2E-5, dC9+TET1+TSDR gRNA=20% avg. suppression, p=0.008, dC9+TET1+CNS1 gRNA=28% avg. suppression, p=3E-5, all vs. dC9+TET1-gRNA) (Figure 1).
Conclusion: Epigenetic editing of FOXP3 using a dCas9-TET1 construct induces DNA demethylation, overexpression, and a regulatory T cell phenotype. Our data are intriguing but need confirmation, particularly to clarify the persistence of induced DNA methylation changes and resistance to phenotype switching. If confirmed, this approach has the potential to significantly improve upon current methods of T-reg generation.
To cite this abstract in AMA style:
Dunn C, Velasco C, Andrews M, Rivas A, Jeffries MA. Epigenetic Editing of FOXP3 in Human T Cells Is Sufficient to Induce Overexpression and Create a Regulatory T Cell Phenotype in Vitro [abstract]. Arthritis Rheumatol. 2018; 70 (suppl 9). https://acrabstracts.org/abstract/epigenetic-editing-of-foxp3-in-human-t-cells-is-sufficient-to-induce-overexpression-and-create-a-regulatory-t-cell-phenotype-in-vitro/. Accessed .« Back to 2018 ACR/ARHP Annual Meeting
ACR Meeting Abstracts - https://acrabstracts.org/abstract/epigenetic-editing-of-foxp3-in-human-t-cells-is-sufficient-to-induce-overexpression-and-create-a-regulatory-t-cell-phenotype-in-vitro/