Session Information
Session Type: Poster Session B
Session Time: 9:00AM-11:00AM
Background/Purpose: The strongest nongenetic risk factors for primary knee OA are advanced age and obesity. We have previously shown a human cartilage microbial DNA signature associated with OA. In this study, we hypothesized that aging and obesity, known to cause gut microbiome shifts, would also produce shifts in cartilage microbial DNA patterns.
Methods: Young (8 week old, n=10) and old (74 week old, n=10) male C57BL/6J mice were divided into equal groups and randomly assigned to chow (NIH-31) or high fat (RD D12492, 60% kcal fat) diet for 8 weeks. Mice were sacrificed at 16 weeks and 82 weeks of age and knee cartilage collected using sterile technique, and DNA extracxted. The V3 and V4 regions of the bacterial rRNA gene were amplified and deep sequenced on an Illumina HiSeq. Operational taxonomic units (OTUs) were assigned in QIIME 1.9.1 using the Greengenes 13_8 97% representative reference set. Group composition differences were compared by Linear Discriminant Analysis Effect Size (LEfSe, LDA-effect sizes ≥ 2 or ≤ -2 were considered significant) following rarefaction.
Results: In both HFD and chow animals, aging was associated with expansion of DNA from members of the phylum Firmicutes (LDA ES 4.8, p=0.009 for chow young vs. old, LDA ES 4.4, p=0.04 for HFD young vs. old, Figure 1A,B), specifically class Clostridia (LDA ES 4.6, p=0.009 for chow, LDA ES 4.3, p=0.03 for HFD, Figure 1A,B) , along with reductions in DNA from members of class Alphaproteobacteria with age (LDA ES 4.3, p=0.02 for chow, LDA ES 3.6, p=0.04 for HFD, Figure 1A,B). Comparing chow to HFD young mice, phylum Proteobacteria was expanded in chow (LDA ES 4.2, p=0.03) whereas members of phylum Firmicutes were expanded in HFD mice (LDA ES 4.3, p=0.02, Figure 2A), specifically including genus Lactococcus (LDA ES 3.7, p=0.005 Figure 2 A,B). Comparing chow to HFD old mice, chow were enriched in order Turicibacterales (LDA ES 4.0, p=0.02, Figure 2A), whereas HFD were enriched in class Thermoleophilia (LDA ES 3.5, p=0.02, Figure 2A) and genus Lactococcus (LDA ES 3.6, p=0.02, Figure 2A,B). We also noted significant increases in the Firmicutes:Bacteroidetes ratio in both HFD and aging (chow young vs. old 2.3±0.7 vs. 29±9, p=0.05, young chow vs HFD 2.3±0.7 vs. 13±3, p=0.04) Figure 1C.
Conclusion: Cartilage microbial DNA patterns vary with nongenetic OA risk factors, including obesity and aging. Previous studies have shown increases in the Firmicutes:Bacteroidetes ratio in the gut microbiome associated with both obesity and aging, we identified a similar pattern within articular cartilage. Future studies should investigate the relationship between cartilage microbial DNA patterns and the gut microbiome, as well as localized innate immune activation driven by intraarticular microbial DNA.
 Microbial clades identified in cartilage of young and old mice fed chow or high-fat diet.
Microbial clades identified in cartilage of young and old mice fed chow or high-fat diet.
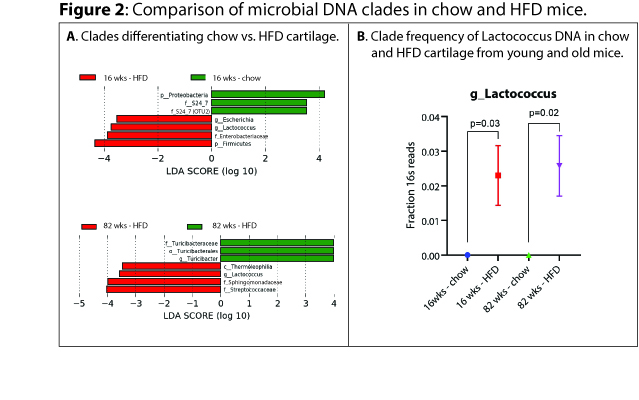 Comparison of microbial clades identified in cartilage of chow-fed and high-fat-diet-fed mice.
Comparison of microbial clades identified in cartilage of chow-fed and high-fat-diet-fed mice.
To cite this abstract in AMA style:
Dunn C, Garman C, Martin J, Izda V, Velasco C, Jeffries M. Distinct Murine Cartilage Microbial DNA Signatures Are Seen in High Fat Diet-Induced Obesity and Aging [abstract]. Arthritis Rheumatol. 2020; 72 (suppl 10). https://acrabstracts.org/abstract/distinct-murine-cartilage-microbial-dna-signatures-are-seen-in-high-fat-diet-induced-obesity-and-aging/. Accessed .« Back to ACR Convergence 2020
ACR Meeting Abstracts - https://acrabstracts.org/abstract/distinct-murine-cartilage-microbial-dna-signatures-are-seen-in-high-fat-diet-induced-obesity-and-aging/
