Session Information
Date: Wednesday, October 24, 2018
Title: 6W024 ACR Abstract: SLE–Etiology & Pathogenesis II (2946–2951)
Session Type: ACR Concurrent Abstract Session
Session Time: 11:00AM-12:30PM
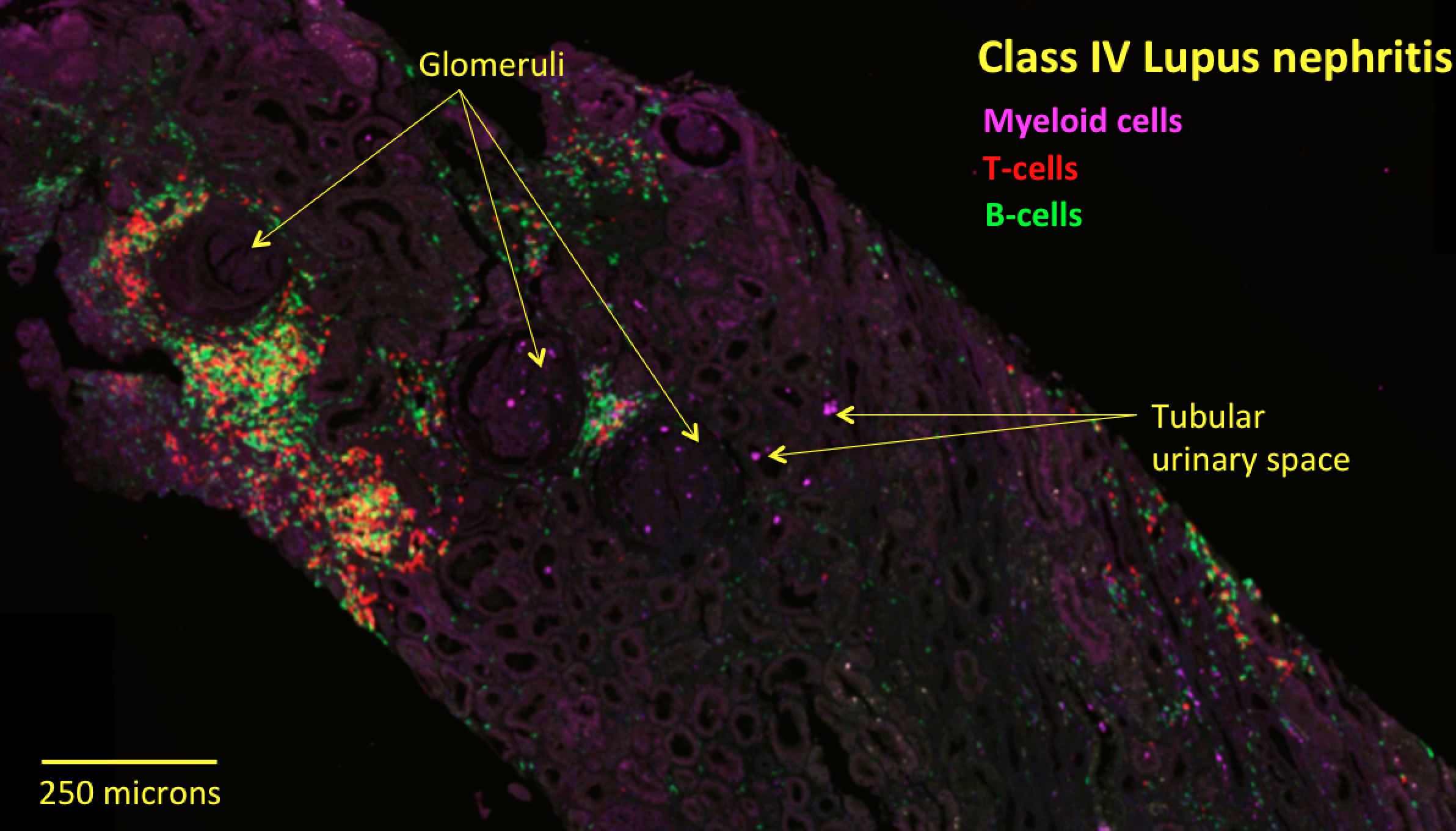
Background/Purpose: The cell-types and pathways driving lupus nephritis (LN) are incompletely understood. The Accelerating Medicine Partnership lupus network Pathway Exploration and Analysis in RenaL disease (AMP-PEARL) consortium sequenced RNA from ~2,900 single cells from LN kidney biopsies from 24 patients and discovered 21 immune cell types, including 5 myeloid subsets. This study provided unprecedented molecular information, but lacked in situ immune cell spatial context and was underpowered for associations with clinical outcomes. We investigated the in situ organization of the immune landscape to determine whether it drives clinical outcomes. Because myeloid cells link innate and adaptive immunity, we hypothesized that their in situ organization reflects the physical configuration of larger immune cell networks that promote kidney remodeling and clinical outcomes.
Methods: We assembled a new cohort of 30 LN patients presenting with Class III or IV disease of varying severity who had their 1st kidney biopsies (na•ve to potent immune-modulators). To dissect the in situ organization of the 5 new myeloid subsets (3 monocyte phenotypes, 1 dendritic cell, and 1 resident macrophage), we converted single cell RNA-sequencing signatures into molecular stains based on highly expressed myeloid subset-specific discriminatory genes with known biological functions. We then stained clinical samples from our cohort for multiplex fluorescent imaging and quantification across tissue sections.
Results: We validated the 5 new myeloid subsets and find they are similarly organized across class III and IV tissue: one monocyte subset (alternatively activated) is excluded from glomeruli but present in the interstitial space; two other monocyte subsets (inflammatory and phagocytic) are present in glomerular and interstitial space and enriched inside tubular urinary space. We also find that DCs are enriched in interstitial over glomerular space.
Conclusion: By converting single cell RNA sequencing information into molecular stains, we developed a novel approach to validate in situ the 5 newly identified myeloid subsets and mapped each to anatomic niches in class III and IV tissue, suggesting molecular organizing principles drive in situ cellular configurations. We are also mapping myeloid subsets in class V LN, histopathological lesions, and examining their connectivity to larger immune cell networks by quantifying the composition of neighboring immune cells (T, B, NK cells) to identify the rules of in situ immune cell organization. We will examine our findings with clinical outcomes, possibly laying the groundwork for disease re-classification based on the immune response and highlighting cell-types and intercellular connections for follow up studies. We anticipate that this approach can be applied to other cell and tissue types.
To cite this abstract in AMA style:
Hoover P, Jones T, Leatherwood C, Waikar S, Costenbader K, Hacohen N. Deconstructing the in Situ Myeloid Cell Microenvironment in Human Lupus Nephritis Tissue [abstract]. Arthritis Rheumatol. 2018; 70 (suppl 9). https://acrabstracts.org/abstract/deconstructing-the-in-situ-myeloid-cell-microenvironment-in-human-lupus-nephritis-tissue/. Accessed .« Back to 2018 ACR/ARHP Annual Meeting
ACR Meeting Abstracts - https://acrabstracts.org/abstract/deconstructing-the-in-situ-myeloid-cell-microenvironment-in-human-lupus-nephritis-tissue/