Session Information
Session Type: Abstract Session
Session Time: 3:00PM-4:30PM
Background/Purpose: Rheumatoid arthritis (RA) is a systemic autoimmune disease with currently no effective prevention strategies. Single-cell technologies have been recently used to investigate established RA heterogeneity, but it is unknown if the immune populations identified from RA tissues are playing important roles in blood during the preclinical phase of disease. Thus, identifying pathogenic immune phenotypes in individuals who can be at risk for future RA, “At-Risk RA”, is crucial to establishing prevention strategies.
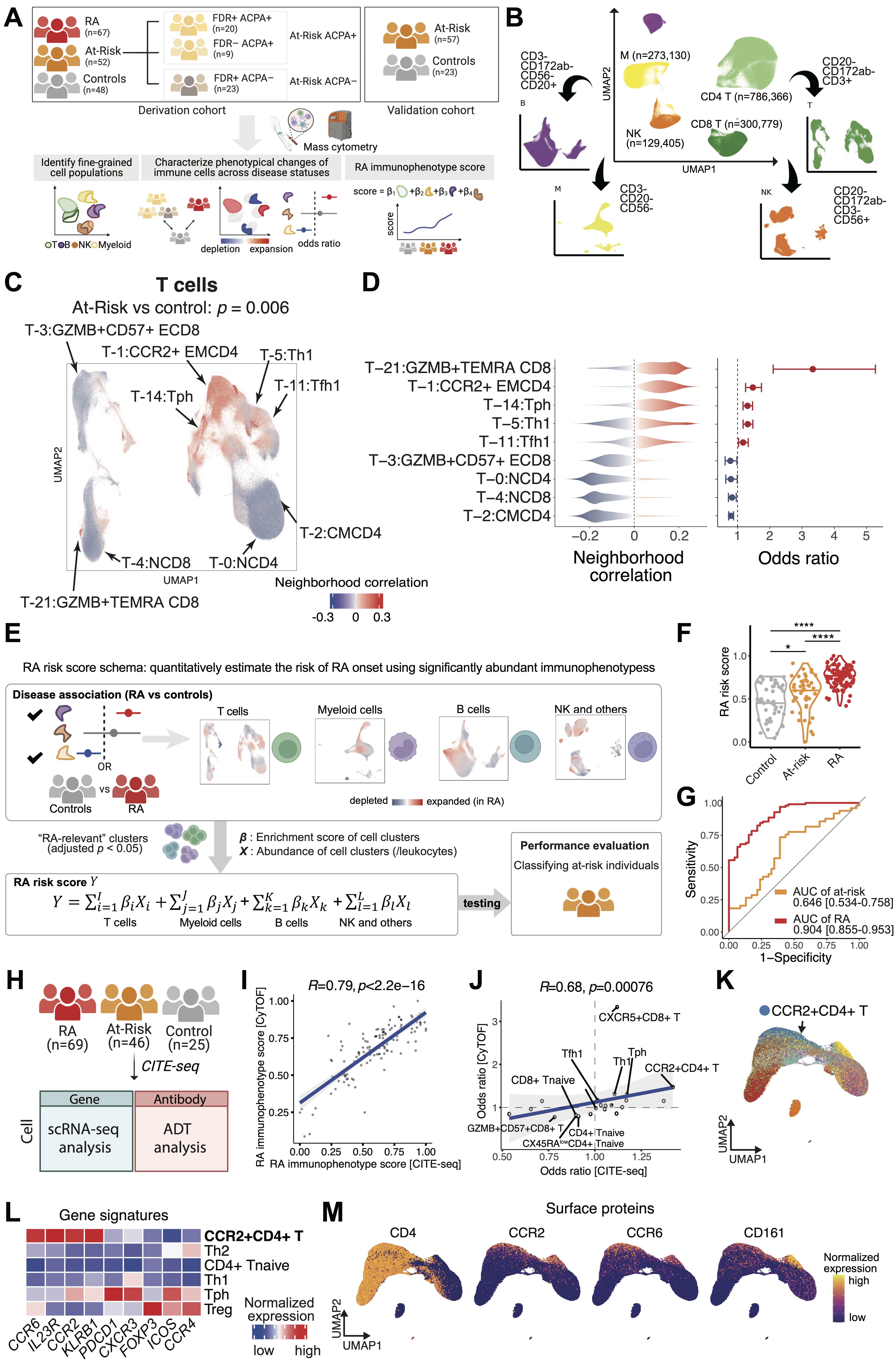
Methods: We applied scalable computational strategies on mass cytometry data to deeply characterize immunophenotypes in blood from At-Risk individuals of clinical subpopulations based on antibodies to citrullinated protein antigens (ACPA) and first-degree relative (FDR) (n=52), and from established RA (n=67), and healthy controls (n=48) (Figure 1A-B). Further, we employed CITE-seq data to blood from At-Risk individuals (n=46), established RA (n=69), and healthy controls (n=25) to validate our findings.
Results: Through integrative and disease association analyses, we quantified the immune populations and uncovered significant cell expansions in At-Risk individuals compared with controls, including CCR2+ T helper cells, T peripheral helper cells (Tphs), type 1 T helper cells, and GZMB+ effector memory T cells that re-express CD45RA (TEMRA) cytotoxic T cells (Figure 1C-D). We further validated the At-Risk associations of these T cell phenotypes using our validation cohort of 57 At-Risk and 23 healthy individuals. In addition, we found that CD15+ classical monocytes were highly expanded in ACPA-negative At-Risk, and an activated PAX5low naïve B cell population was expanded in ACPA-positive individuals who also had an FDR with RA. Further, we developed a “RA immunophenotype score” classification method based on the degree of enrichment and the abundance of cell states relevant to established RA using mixed-effect modeling and logistic regression (Figure 1E). We found this score significantly distinguished At-Risk individuals from the controls (p=0.039 and AUC >0.6) (Figure 1F-G). In CITE-seq data (Figure 1H), we observed significant correlation of both RA immunophenotype scores (Figure 1I) and At-Risk-association effect size of T cell subsets (Figure 1J) between those derived from CyTOF data and those obtained from CITE-seq data, reinforcing the robustness and reliability of our scoring method across diverse technological platforms. Finally, we discovered high expression of Th17-related genes in CCR2+CD4+ T cells in CITE-seq data (Figure 1K-M).
Conclusion: We systematically characterized altered circulating immune phenotypes in At-Risk individuals, and immunophenotypical differences among ACPA+ and FDR At-Risk subpopulations. Our classification model may provide a promising approach to understand the pathogenesis of RA with the goal to developing preventive strategies.
To cite this abstract in AMA style:
Inamo J, Keegan J, Griffith A, Ghosh T, Horisberger A, Howard K, Pulford J, Murzin E, Hancock B, Eisenhaure T, Dominguez S, Gurra M, Gurajala S, Jonsson A, Seifert J, Feser M, Norris J, Cao Y, Apruzzese W, Bridges S, Bykerk V, Goodman S, Donlin L, Firestein G, Bathon J, Hughes L, Tabechian D, Filer A, Pitzalis C, Anolik J, Moreland L, Hacohen N, Guthridge J, James J, Cuda C, Perlman H, Brenner M, Raychaudhuri S, Sparks J, Holers M, Deane K, Lederer J, Rao D, Zhang F. Deciphering Pathogenic Phenotypes by Multi-modal Deep Single-cell Blood Immunophenotyping in Individuals At-risk for Rheumatoid Arthritis [abstract]. Arthritis Rheumatol. 2024; 76 (suppl 9). https://acrabstracts.org/abstract/deciphering-pathogenic-phenotypes-by-multi-modal-deep-single-cell-blood-immunophenotyping-in-individuals-at-risk-for-rheumatoid-arthritis/. Accessed .« Back to ACR Convergence 2024
ACR Meeting Abstracts - https://acrabstracts.org/abstract/deciphering-pathogenic-phenotypes-by-multi-modal-deep-single-cell-blood-immunophenotyping-in-individuals-at-risk-for-rheumatoid-arthritis/