Session Information
Session Type: Poster Session A
Session Time: 10:30AM-12:30PM
Background/Purpose: Transcriptional profiling represents a powerful tool for deciphering disease-relevant pathways and deriving functional biomarkers. Given the shared demographic, genetic, and clinico-epidemiologic features between rheumatoid arthritis-associated interstitial lung disease (RA-ILD) and idiopathic pulmonary fibrosis (IPF), we sought to demonstrate the mechanistic basis for similarities (as well as differences) between RA-ILD and IPF through comparative single cell RNA sequencing (scRNAseq) of lung tissue from these disorders.
Methods: We obtained explanted lung tissue from n=13 IPF and n=3 RA-ILD patients as well as n=10 controls (source of healthy lungs rejected for transplantation). Following single cell isolation, RNA extraction, and cDNA amplification, we generated gene expression libraries from individual lung samples and then performed scRNAseq using the 10x Genomics’ platform. Resulting sequences were aligned with Cellranger and clustered using the Seurat R toolkit. This approach yielded differentially expressed genes (DEGs) in specific cell populations derived from IPF, RA-ILD, and Control lung tissue. Gene Ontology algorithms were used for analysis of distinguishing as well as overlapping pathways represented by these DEGs.
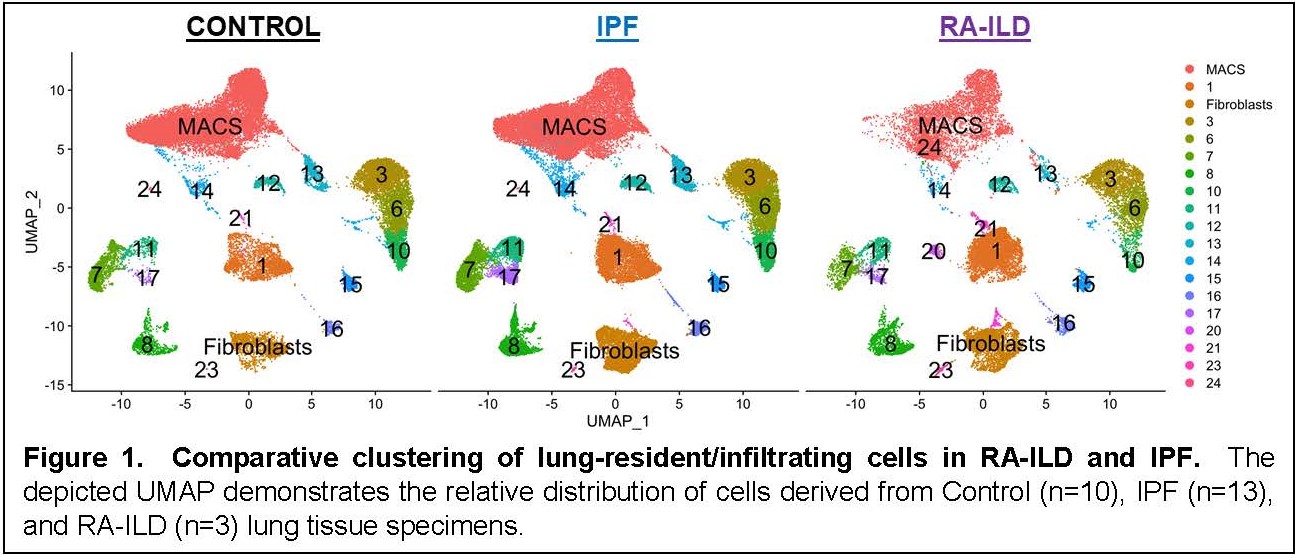
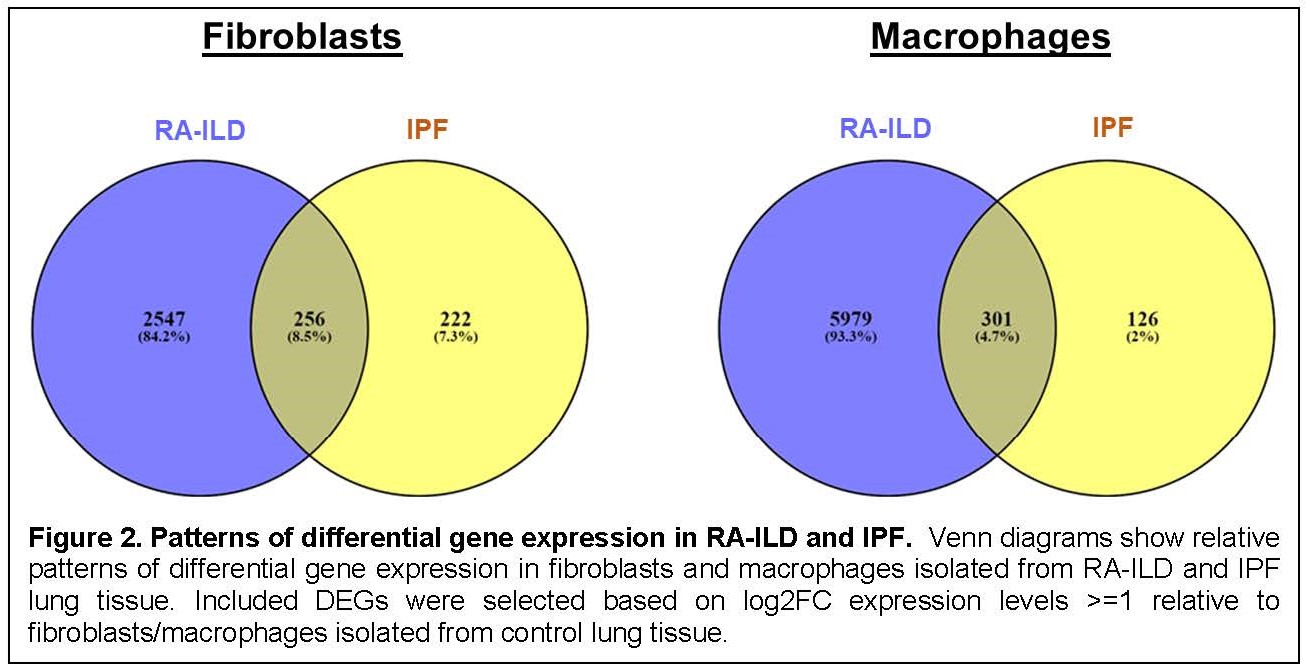
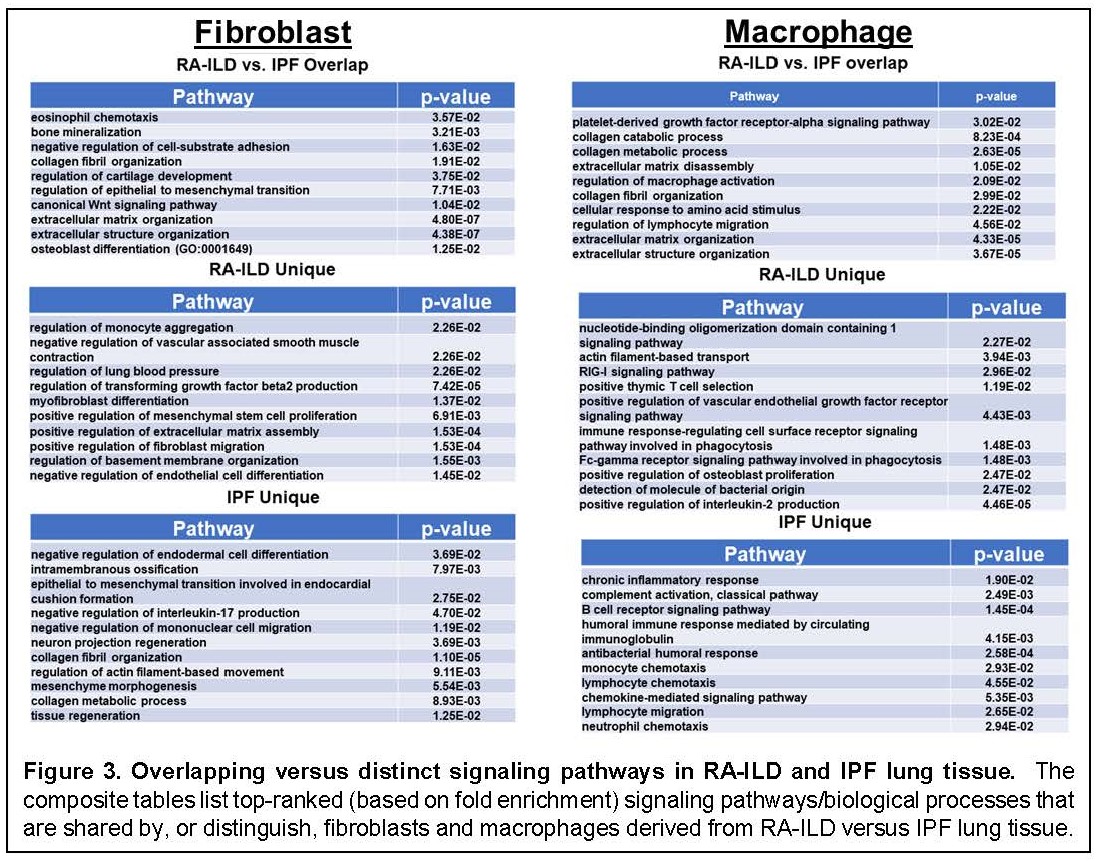
Results: Clustering demonstrated prominent macrophage and fibroblast populations in RA-ILD, IPF, and control lung tissue (Figure 1). Assessment of DEGs in these cell populations (each comprised of multiple subclusters) revealed numerous DEGs separating both IPF and RA-ILD from control lung specimens. Comparison of these macrophage- and fibroblast-derived DEGs between RA-ILD and IPF yielded a series of overlapping as well as distinct DEGs in both macrophages and fibroblasts (Figure 2). Although there were a significantly greater number of DEGs specifically associated with macrophages and fibroblasts derived from RA-ILD lung tissue (compared to IPF), a considerable percentage of IPF-associated DEGs were also represented in RA-ILD cell populations [256/478 (54%) IPF-associated macrophage DEGs and 301/426 (71%) IPF-associated fibroblast DEGs matched with RA-ILD DEGs]—highlighting the degree of transcriptional overlap between these disorders. Key pathways derived from Gene Ontology analysis of overlapping as well as unique DEGs in RA-ILD and IPF lung tissue are shown in Figure 3. While many of the shared DEGs represent pathways geared toward tissue remodeling and fibrosis, those DEGs distinguishing RA-ILD from IPF feature a range of both pro-inflammatory (macrophage-oriented) and pro-fibrotic (fibroblast-directed) pathways.
Conclusion: Assessment of transcriptional profiles linked to RA-ILD and IPF lung tissue demonstrates both shared and unique patterns of gene expression in these disorders. Corresponding Gene Ontology analysis highlights key pathways encompassing mediators that a) are likely to play a role in disease pathogenesis and b) should provide a template for derivation of peripheral blood biomarkers as well as selection of therapeutic targets.
To cite this abstract in AMA style:
Sciurba J, Kass D, Valenzi E, Sembrat J, Lafyatis R, Ascherman D. Comparative Transcriptional Profiling of IPF and RA-ILD Lung Tissue Demonstrates Both Overlapping and Distinct Cell-specific Signaling Pathways [abstract]. Arthritis Rheumatol. 2024; 76 (suppl 9). https://acrabstracts.org/abstract/comparative-transcriptional-profiling-of-ipf-and-ra-ild-lung-tissue-demonstrates-both-overlapping-and-distinct-cell-specific-signaling-pathways/. Accessed .« Back to ACR Convergence 2024
ACR Meeting Abstracts - https://acrabstracts.org/abstract/comparative-transcriptional-profiling-of-ipf-and-ra-ild-lung-tissue-demonstrates-both-overlapping-and-distinct-cell-specific-signaling-pathways/