Session Information
Session Type: Abstract Submissions (ACR)
Background/Purpose: Cartilage secretome contains a complex matrix of proteolytically derived peptides that may provide useful information of the biological events occurring in the articular joint in normal and pathological condition. Moreover, the low-molecular-weight subset of the cartilage proteome may have a diagnostic potential in osteoarthritis (OA) research, not fully exploited yet. The aim of this study is to investigate cartilage secretome by means of peptidome analysis and to provide a novel source for OA biomarker discovery.
Methods: Tissue explants were obtained from the dissection of human OA cartilages, both from the wounded zone (WZ) and the unwounded zone (UZ). The study was approved by the local ethical committee. Cartilage shavings from each zone were cut into 6 mm discs and five discs/zone were placed into 96 wells plates and incubated (37 °C/5% CO2) in serum-free DMEM up to 6 days. Conditioned media from each condition were collected at day 1, 3 and 6. Selective extraction of endogenous peptides was realized by combining ultrafiltration and solid phase extraction. Then, the peptides were directly analyzed by nano reversed-phase liquid chromatography (RP-LC) in an Easy nLC II system coupled to an ion trap LTQ Orbitrap Velos Pro mass spectrometer (Thermo Scientific) without trypsin digestion. Protein/peptide identifications were searched against human database using the SEQUEST algorithm (Proteome Discoverer 1.3, Thermo Scientific).
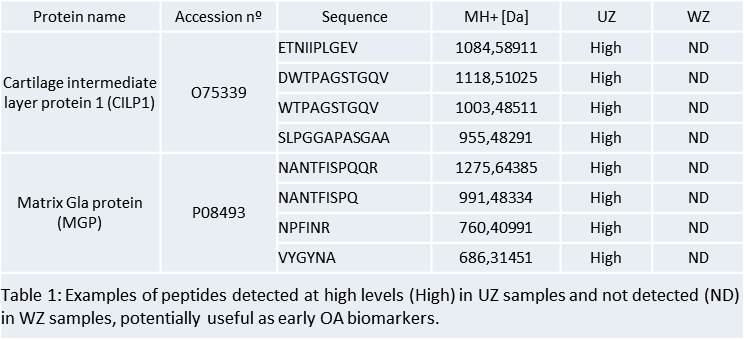
Results: Firstly we compared the profiles of peptides released to the media at different times (0, 3, and 6 days in culture). At day 3 we detected the highest number of unique peptides and proteins, and the lowest serum contamination. Thus, we selected day 3 as the best point for the next proteomic analysis in which we compared peptide profiles from WZ and UZ. This study led to the detection of a panel of 262 peptides corresponding to 36 proteins that were differentially released from the WZ and UZ in OA cartilage. Most of them were cartilage ECM proteins or proteins with well-established matrix functions, such as collagens and proteoglycans, thus indicating the high cartilage-specific biomarker concentration present in our samples. 18 peptides were detected only in the WZ (corresponding to 5 proteins: K1C9, K1C10, PRPC, FIBA, and DCD) while 60 peptides only in the UZ (corresponding to 20 proteins: ACAN, CILP1, CILP2, COL11A2, COL1A2, COL25A1, LTBP2, MGP, and PRELP among others) (Table 1). 11 proteins (BGN, CLU, COL1A1, COL2A1, COL3A1, COL5A2, COMP, and FN1 among others) were detected in both zones but in all the cases the number of peptides was higher in the UZ than in the WZ.
Conclusion: To our knowledge this is the first proteomic study specifically focused on cartilage peptidome analysis. In this work we analysed the cartilage endogenous peptides profile and highlighted its potential application as novel OA biomarker source for early disease detection.
Disclosure:
V. Calamia,
None;
L. Lourido,
None;
D. López,
None;
J. Mateos,
None;
P. Fernández-Puente,
None;
C. Fernández-Costa,
None;
B. Rocha,
None;
C. Ruiz-Romero,
None;
F. J. Blanco,
None.
« Back to 2013 ACR/ARHP Annual Meeting
ACR Meeting Abstracts - https://acrabstracts.org/abstract/cartilage-peptide-profiling-identifying-novel-oa-biomarkers-for-early-disease-detection/