Session Information
Session Type: Poster Session B
Session Time: 10:30AM-12:30PM
Background/Purpose: The minor allele of the common rs2231142 (Q141K) ABCG2 variant predicts inadequate response to allopurinol urate lowering therapy (ULT). We hypothesize that additional variants in genes encoding urate secretory and reuptake transporters and allopurinol-to-oxypurinol metabolic enzymes also predict allopurinol response.
Methods: This study included a subset of participants with gout from the Long-term Allopurinol Safety Study Evaluating Outcomes in Gout Patients trial (NCT01391325), whose whole genome was sequenced (n=667). Participant exclusion steps and individual-level quality control filtering of sequencing data left 563 of 667 participants. Good responders had ≥1 good response visits (serum urate (SU) < 0.36mmol/L on allopurinol ≤300 mg/day) and no poor response visits (SU ≥0.36mmol/L despite allopurinol >300 mg/day) or a 5:1 ratio of good to poor responses over 5 to 6 monthly timepoints. Inadequate responders had no good response visits and ≥1 poor response visits, or a 1:5 ratio of good to poor responses. Adherence to allopurinol was determined by pill counts, and for a subgroup (n = 303), by plasma oxypurinol >20μmol/l. Using the sequence kernel association test (SKAT) we estimated the combined effect of rare and common protein-coding variants in urate secretory genes (ABCC4, ABCC5, ABCG2, SLC17A1, SLC17A3, SLC22A6, SLC22A8), urate reuptake genes (SLC2A9, SLC22A11) and allopurinol-to-oxypurinol metabolic genes (AOX1, MOCOS, XDH) on allopurinol response.
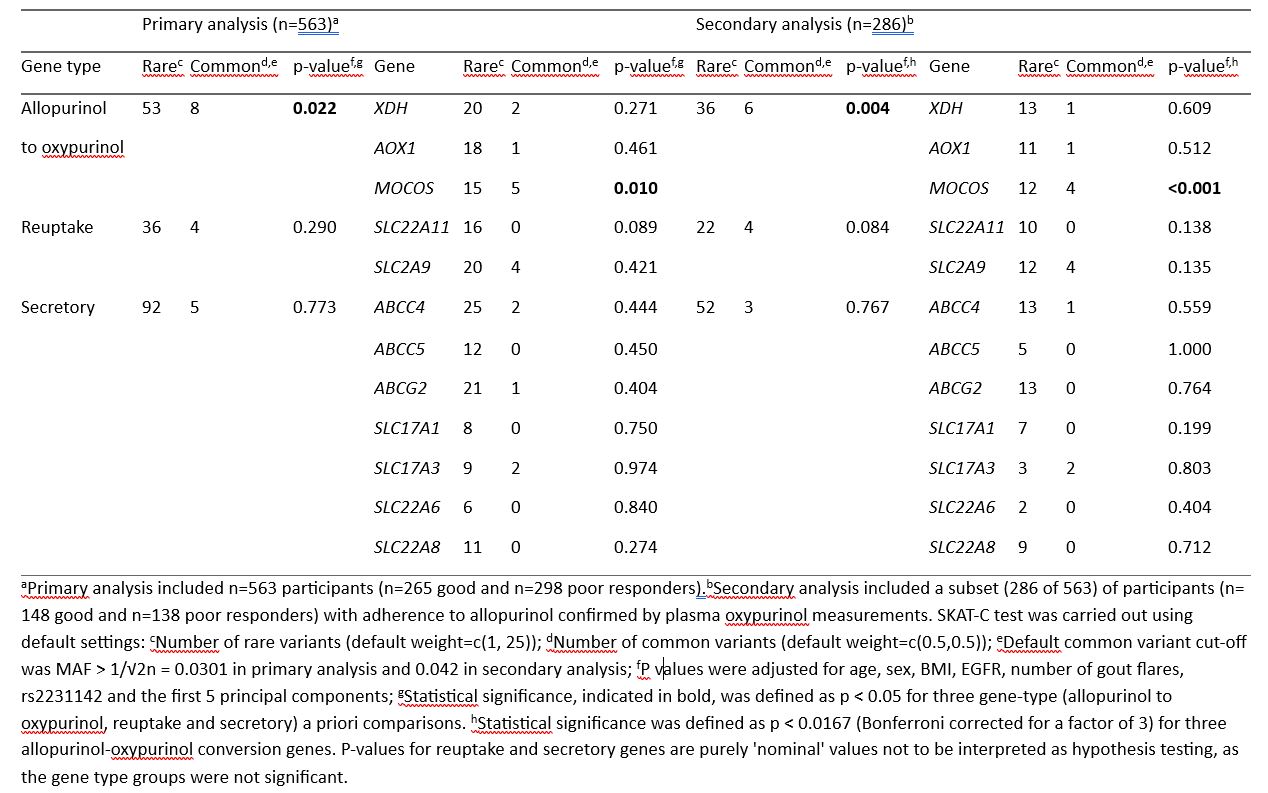
Results: There was an association of all rare and common protein-coding variants in the allopurinol-to-oxypurinol gene group (PSKAT‑C = 0.022), but not in the reuptake (PSKAT C = 0.29) or secretory (PSKAT C = 0.77) gene groups, with allopurinol response (Table 1). Pooled rare and common protein-coding variants in the allopurinol-to-oxypurinol metabolism gene MOCOS, encoding molybdenum cofactor sulphurase were associated with allopurinol response (PSKAT‑C = 0.010), while variants in other allopurinol metabolic genes tested, AOX1 (PSKAT‑C = 0.46) or XDH (PSKAT‑C = 0.27) were not (Table 1). Evidence for genetic association with allopurinol response in the allopurinol-to-oxypurinol gene group (PSKAT‑C = 0.004) and MOCOS (PSKAT‑C < 0.001) was stronger when adherence to allopurinol therapy was confirmed by measuring plasma oxypurinol (Table 1). The association of MOCOS with response was independent of age, sex, BMI, eGFR, number of gout flares in the past year, ABCG2 rs2231142, pre-ULT serum urate measures and the top 5 genetic principal components.
Conclusion: We provide evidence for common and rare genetic variation in MOCOS associating with allopurinol response. The statistical strength of the signal was improved with more accurate phenotyping of adherence to allopurinol therapy. These results support the idea that additional genetic factors play a part in determining response to allopurinol and point to a mechanism involving allopurinol to oxypurinol conversion.
To cite this abstract in AMA style:
Fanning N, Cadzow M, Topless R, Frampton C, Dalbeth N, Merriman T, Stamp L. Association of Rare and Common Genetic Variants in MOCOS with Inadequate Response to Allopurinol [abstract]. Arthritis Rheumatol. 2024; 76 (suppl 9). https://acrabstracts.org/abstract/association-of-rare-and-common-genetic-variants-in-mocos-with-inadequate-response-to-allopurinol/. Accessed .« Back to ACR Convergence 2024
ACR Meeting Abstracts - https://acrabstracts.org/abstract/association-of-rare-and-common-genetic-variants-in-mocos-with-inadequate-response-to-allopurinol/