Session Information
Session Type: Poster Session A
Session Time: 9:00AM-11:00AM
Background/Purpose: SLE is a chronic and debilitating disease; in the USA with an estimated incidence of 3-10 per 100,000 people and currently affecting an estimated 300,000 people. The exact pathogenesis of SLE remains unknown, but there is a growing body of evidence that various genetic, hormonal, and environmental factors contribute to its occurrence and severity. Each is thought to modulate the activation and regulation of innate and adaptive immunity in ways that drive autoimmunity, the underlying cause of SLE. The hallmark of autoimmunity in SLE is B cell hyperactivity leading to the generation of autoantibodies, which together drives the deposition of deleterious immune complexes. SLE has strong familial clustering, high heritability, and high concordance rates between monozygotic versus dizygotic twins, suggesting it’s a polygenic disease. Among the many different gene variants reportedly associated with SLE risk or severity are single nucleotide polymorphisms (SNPs) in the integrin alpha-M (ITGAM) gene, which are present in more than 1/3 of SLE patients. ITGAM encodes the protein CD11b, which pairs with CD18 and forms the Mac-1 receptor. Among the ITGAM SNPs associated to SLE is SNP rs1143678, encoding a P1146S amino acid substitution in the cytoplasmic tail of CD11b. Mac-1 is widely expressed by myeloid cells, and it is reported that SNP rs1143678 alters myeloid cell functions in ways that could propel SLE. Mac-1 is also expressed on lymphocytes, but comparatively little is known about its role in B cells or whether SNP rs1143678 has any impact on B cell biology. To fill this gap in knowledge we are investigating the impact of ITGAM SNP rs1143678 on Epstein-Barr Virus (EBV) transformed B cells from healthy human donors.
Methods: N/A
Results: We report here three of our main findings. First, juxtaposition of the cytoplasmic tails of CD11b and CD18 – and their activation induced separation – are thought to be required for Mac-1-mediated cell signaling. Our in silico structural modelling (Fig. 1) predicts that the SLE-associated P1146S substitution significantly changes the conformation of CD11b’s cytoplasmic tail. This shape-shift should allow CD11b to accommodate additional or new cytoskeletal/cytoplasmic binding partners, and thereby could influence Mac-1 signaling. Second, phosphorylation of 1142Ser (an invariant residue in CD11b) is thought to be required for Mac-1 signaling, but using in vitro kinase assays and Western blots we provide the first direct evidence that the SLE-associated 1146Ser residue is also phosphorylated (data not shown). Third, by mass-spectrometry combined with proteomics and network analyses, we obtained evidence that both IFNγ and antigen presentation pathways are enriched in B cells from donors expressing the ITGAM SNP variant (Fig 2). Indeed, compared to B cells from ITGAM invariant donors, B-cells from donors carrying the ITGAM SNP showed increased expression of CD11b, increased spontaneous secretion of IFNγ, and increased expression of the IFNγR2 receptor following stimulation (Fig 3). This trio of new findings together suggests that the ITGAM SNP rs1143678 significantly alters the biology of B cells, which might explain the strong association rs1143678 to SLE.v
Conclusion: N/A
 Fig 1. Space filling (top) and ribbon models (bottom) of the cytoplasmic tail of CD11b, viewed from inside the cell looking upwards at the receptor. For individuals carrying the SNP rs1143678 (right) the modelling predicts a 12-angstrom-wide space between the invariant 1142SER and the SNP variant 1146Ser residues (dashed yellow line).
Fig 1. Space filling (top) and ribbon models (bottom) of the cytoplasmic tail of CD11b, viewed from inside the cell looking upwards at the receptor. For individuals carrying the SNP rs1143678 (right) the modelling predicts a 12-angstrom-wide space between the invariant 1142SER and the SNP variant 1146Ser residues (dashed yellow line).
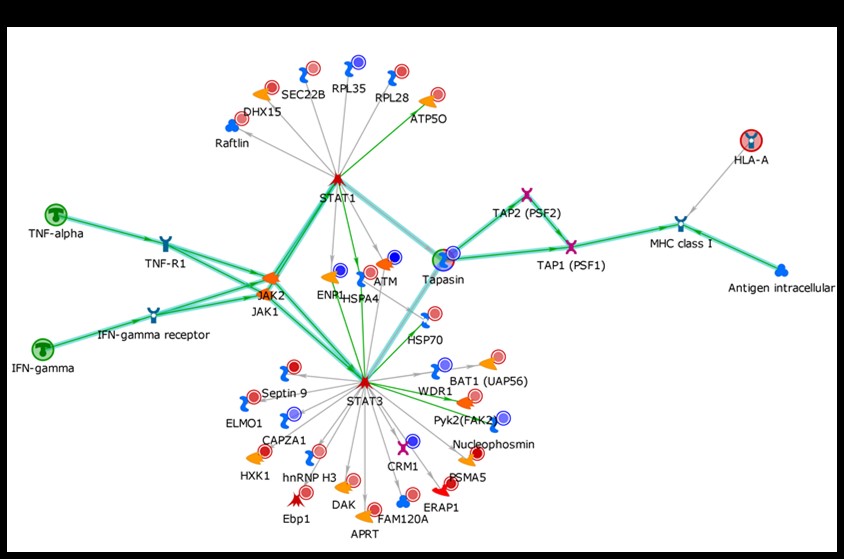 Fig 2. B cells from healthy donors homozygous for the ITGAM SNP rs1143678 or the major allele were transformed with Epstein Bar Virus. The cells were lysed and whole proteome analysis was done by LC MS/MS. Protein expression differences were determined and over-represented pathways in the SNP samples were mapped by Clarivate Analytics pathway analysis software. B-cells expressing the ITGAM SNP rs1143678 had high levels of IFNg induced MHC-1 presentation.
Fig 2. B cells from healthy donors homozygous for the ITGAM SNP rs1143678 or the major allele were transformed with Epstein Bar Virus. The cells were lysed and whole proteome analysis was done by LC MS/MS. Protein expression differences were determined and over-represented pathways in the SNP samples were mapped by Clarivate Analytics pathway analysis software. B-cells expressing the ITGAM SNP rs1143678 had high levels of IFNg induced MHC-1 presentation.
 Fig 3. A) B-cells expressing the SLE associated SNP have a higher basal expression of CD11b. B) B-cells expressing the SLE associated SNP spontaneously secrete higher amounts of IFNg. C) Upon stimulation with exogenous IFNg, B-cells expressing the SLE associated SNP increase their surface expression of IFNgR2 to a higher degree than contol. A * indicates p < 0.05 by T test.
Fig 3. A) B-cells expressing the SLE associated SNP have a higher basal expression of CD11b. B) B-cells expressing the SLE associated SNP spontaneously secrete higher amounts of IFNg. C) Upon stimulation with exogenous IFNg, B-cells expressing the SLE associated SNP increase their surface expression of IFNgR2 to a higher degree than contol. A * indicates p < 0.05 by T test.
To cite this abstract in AMA style:
Blake J, Szalai A, Edberg J, Mobley J. An SLE-linked ITGAM Gene Variant Changes Mac-1 Structure, Signaling, and Surface Expression and Enhances IFNg Production and Antigen Presentation by B Cells [abstract]. Arthritis Rheumatol. 2020; 72 (suppl 10). https://acrabstracts.org/abstract/an-sle-linked-itgam-gene-variant-changes-mac-1-structure-signaling-and-surface-expression-and-enhances-ifng-production-and-antigen-presentation-by-b-cells/. Accessed .« Back to ACR Convergence 2020
ACR Meeting Abstracts - https://acrabstracts.org/abstract/an-sle-linked-itgam-gene-variant-changes-mac-1-structure-signaling-and-surface-expression-and-enhances-ifng-production-and-antigen-presentation-by-b-cells/
