Session Information
Session Type: Abstract Session
Session Time: 11:30AM-11:45AM
Background/Purpose: We discovered 21 immune cell-types in lupus nephritis kidney biopsies as part of the Accelerating Medicines Partnership (AMP) consortium. These immune cells are the basis of new hypotheses about the drivers of disease. However, we cannot feasibly collect immune cells from human kidney biopsies for mechanistic testing. Mouse lupus models and humans share important clinical features including autoantibody development and kidney injury that progresses to failure. How mice recapitulate human lupus nephritis remains an open question. Here, we used single cell RNA sequencing to identify homologous intrarenal immune cell subsets and conserved gene programs from humans and four common lupus strains.
Methods: Using AMP protocols we sorted CD45+ cells from dissociated mouse kidneys with spontaneous adaptive-driven autoimmunity (NZB/W and MRL/lpr) and TLR7-overexpression innate-driven autoimmunity (Sle1.Yaa and NZW/BXSB) in early and nephritic disease. We profiled single cell transcriptomes using 10x Genomics and analyzed droplets that contained >500 genes and UMIs after doublet removal using Seurat 3.0. We analyzed the top 2000 variable genes for clustering and differential gene expression. We compared transcriptomes of intrarenal immune cells collected from mice at early and peak clinical disease to those from humans at peak clinical disease.
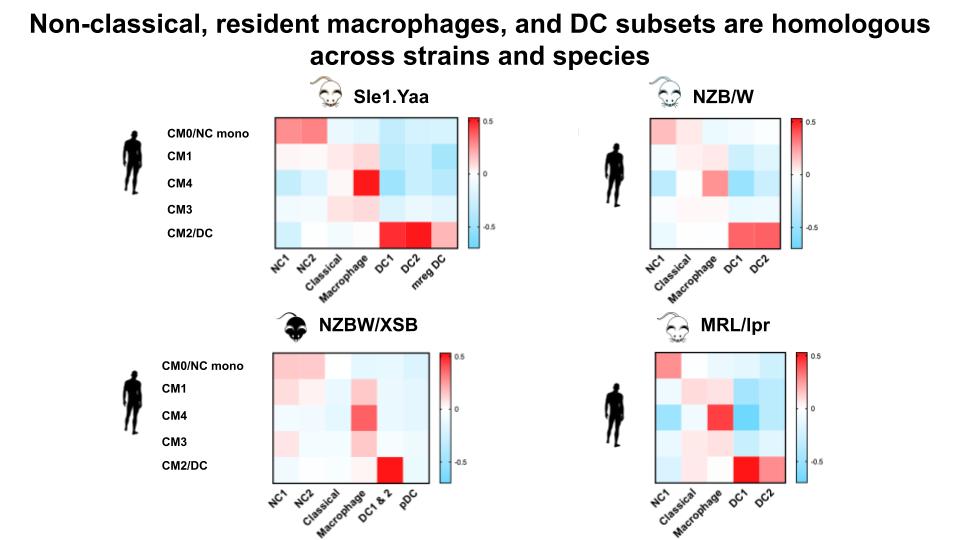
Results: We identified intrarenal myeloid, T-, and B-cell subsets from >75,000 cells passing QC. Many subsets were shared among strains and homologous to human subsets. In particular, the myeloid compartment in mice and humans contained: i) non-classical monocytes that expressed TNF and fate related transcription factors (Spi1, Nr4a1); ii) a novel Fcrl5+ non-classical monocyte unique to TLR7-overexpression strains that emerged in nephritic mouse kidneys and expressed Nr1h3 known to regulate lipids and inflammation; iii) resident-like macrophages that expressed genes for tissue repair (Igf1, Pdgfrb), immunomodulation (Creb5, Dab2), and phagocytosis (Mertk, Gas6); resident-like macrophages in adaptive-driven lupus strains shifted gene expression from anti-fibrotic to remodeling that correlated with proteinuria. We spatially mapped these homologous cells in kidney sections and found they localized to similar compartments in mice and in most human patients.
Conclusion: Most intrarenal myeloid, T-, and B-cell subsets in early and nephritic disease were common to mouse strains; unique subsets correlated with genetic susceptibility. TLR7-overexpression strains contained a novel and unique Fcrl5+ non-classical monocyte. Spontaneous strains contained a population of residential-like macrophages that shifted gene expression from anti-fibrotic to remodeling in nephritic mice. Both non-classical and residential-like macrophages in mice and humans shared genes critical for tissue repair, immunomodulation, and immune cell recruitment. Further, each of these cell-types localized to similar locations in kidney sections in mice and a subset of human patients. These findings support shared roles of these cells in lupus nephritis. We hope our work opens a new path using mouse models to more precisely study aspects of human disease.
To cite this abstract in AMA style:
Hoover P, Peters M, Lieb D, Wang R, Dunlap G, Rao D, Hacohen N, Davidson A. An Atlas of Human and Mouse Intrarenal Immune Cells in Lupus Nephritis Reveals Homologous Immune Populations Across Common Mouse Strains and Species [abstract]. Arthritis Rheumatol. 2021; 73 (suppl 9). https://acrabstracts.org/abstract/an-atlas-of-human-and-mouse-intrarenal-immune-cells-in-lupus-nephritis-reveals-homologous-immune-populations-across-common-mouse-strains-and-species/. Accessed .« Back to ACR Convergence 2021
ACR Meeting Abstracts - https://acrabstracts.org/abstract/an-atlas-of-human-and-mouse-intrarenal-immune-cells-in-lupus-nephritis-reveals-homologous-immune-populations-across-common-mouse-strains-and-species/