Session Information
Session Type: Poster Session A
Session Time: 9:00AM-11:00AM
Background/Purpose: Alternative splicing of mRNA results in important biological impacts with increasing evidence implicating it in the pathology of autoimmune diseases. However, it is understudied in idiopathic inflammatory myopathies (IIM).
Methods: We studied alternative splicing in 152 muscle biopsies from IIM patients and controls. Patients included subgroups with myositis-specific autoantibodies including (anti-Jo1, anti-HMGCR, anti-SRP, anti-NXP2, anti-TIF1g, anti-Mi2, anti-MDA5), and inclusion body myositis (IBM). RNA sequencing data were aligned to the reference genome (GRCh38.p13), then alternative splicing events were quantified using SplAdder. Expected protein changes were identified and visualized using RiboSplitter. The percent spliced-in (PSI) was calculated as reads supporting isoform 2 divided by total reads supporting either isoform per event. We used an alpha of 0.05 and adjusted p values for multiple comparisons. Statistically significant events were filtered based on magnitude of difference to detect events more likely to be biologically meaningful. Finally, we identified outlier splicing events with PSI difference of 3 times the interquartile range, when compared to all other IIM and control samples.
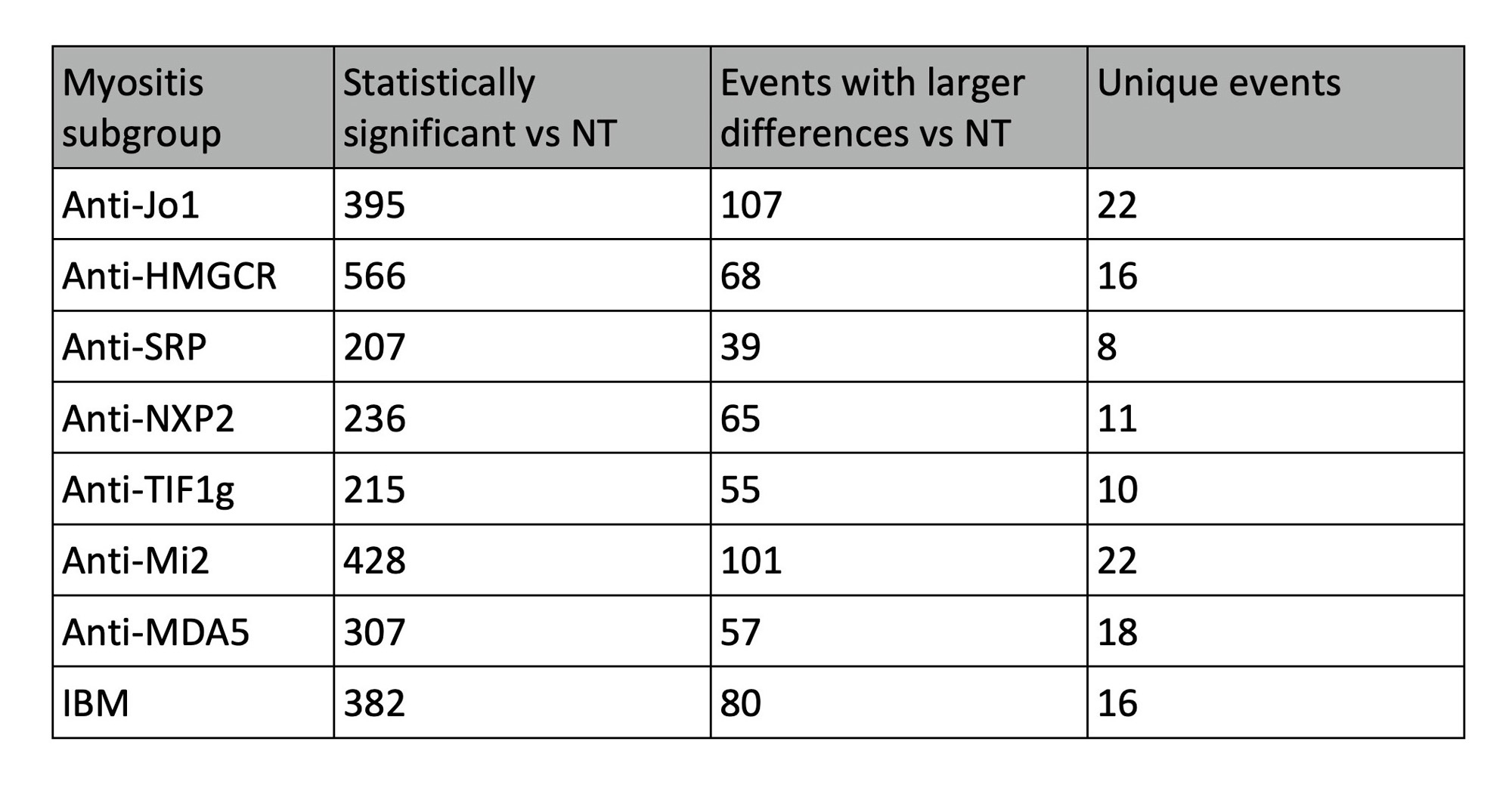
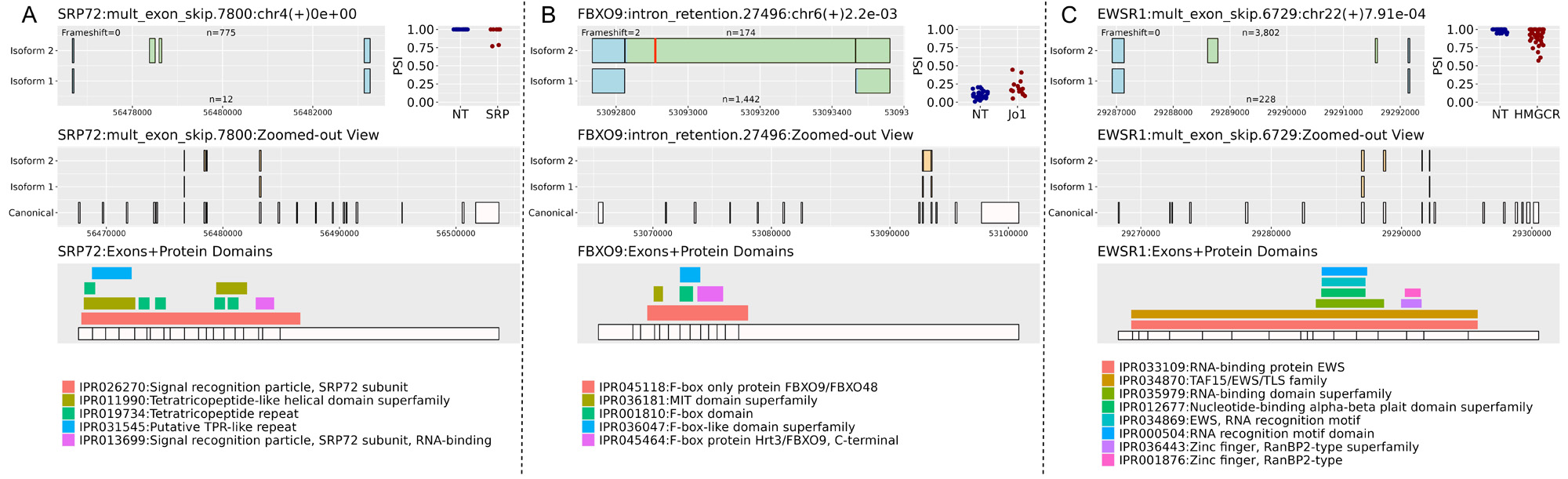
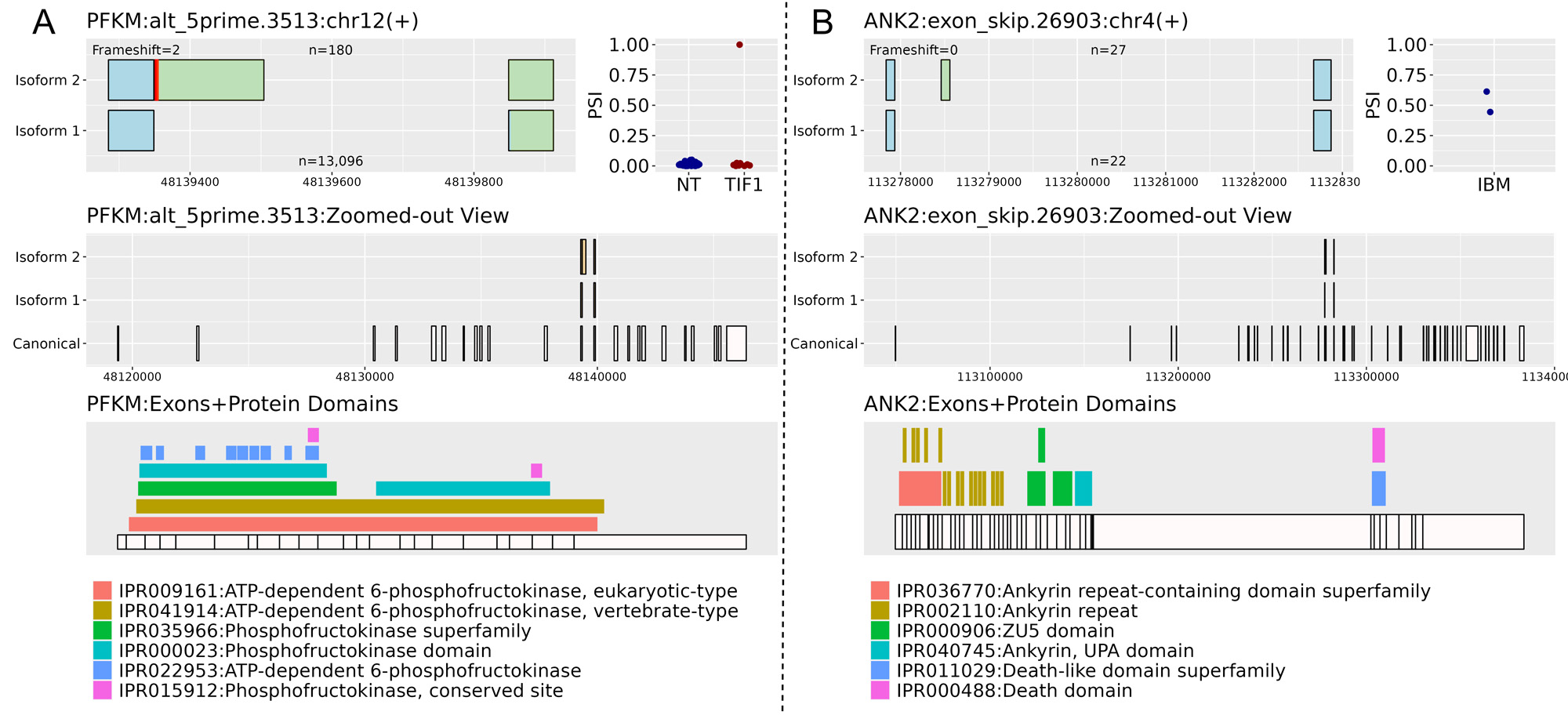
Results: There were hundreds of statistically significant alternative splicing events in each IIM subgroup vs controls. Overall, there were 337 events (in 265 protein coding genes) with larger delta PSI. For each subgroup, we subtracted events that were also present in other subgroup comparisons, resulting in a smaller list of altered splicing that is unique to each subgroup (Table 1). We discovered unique splicing in two patients with anti-SRP myositis where exons 7 and 8 of SRP72 were skipped leading to loss of a tetratricopeptide repeat region (Fig 1A) that is involved in protein binding. SRP72 encodes one of the 6 protein subunits of the signal recognition particle (SRP). An intron retention event in the anti-Jo1 subgroup introduced a stop codon after exon 9 of FBXO9 that would result in a truncated protein (Fig 1B). Patients with HMGCR were more likely to express isoform 1 of an event in EWS RNA binding protein 1, skipping exons 8 and 9 (Fig 1C). One patient with TIF1g, and two with NXP2 had 49, 26, and 10 outlier events, respectively. One of the TIF1g outlier events was an alternative 5´event in the muscle-specific phosphofructokinase gene resulting in a longer 12th exon with introduction of a stop codon that would disrupt the C-terminal phosphofructokinase domain and conserved site (Fig 2A). Additionally, there was a single event that only appeared in two patients with IBM where the 15th exon of ankyrin 2 was skipped, which corresponds to an ankyrin repeat domain (Fig 2B).
Conclusion: We present results of a systematic unbiased search of alternative splicing in IIM muscles. Splicing events that are unique to a myositis subgroup are more likely to be relevant to disease pathology since they were not differentially present in any other subgroup. We discovered a unique event in SRP72 in the anti-SRP subgroup, connecting a genomic feature to the disease’s specific autoantibody. A few patients expressed roughly two thirds of the total outlier events, suggesting generally perturbed splicing in these patients.
NT= Normal tissue
Top left figure shows a zoomed-in view of the alternative splicing event. The title includes gene name, event ID, chromosome, strand, and adjusted p value. n is the number of reads supporting isoform 1 or 2. Red lines represent the first stop codon per isoform. Light blue exons indicate same protein product, while light green indicate altered peptides when translated.
Top right figure shows percent-spliced in (PSI) of isoform 2 by myositis subgroup and normal tissue (NT).
Middle figure shows a zoomed-out view of the alternative splicing event with complete exons of the canonical transcript.
Bottom figure shows exons aligned with protein domains of the gene.
Panels A, B, and C illustrate three events in SRP, Jo1, and HMGCR subgroups, respectively
Top left figure shows a zoomed-in view of the alternative splicing event. The title includes gene name, event ID, chromosome, strand, and adjusted p value. n is the number of reads supporting isoform 1 or 2. Red lines represent the first stop codon per isoform. Light blue exons indicate same protein product, while light green indicate altered peptides when translated.
Top right figure shows percent-spliced in (PSI) of isoform 2 by myositis subgroup and normal tissue (NT).
Middle figure shows a zoomed-out view of the alternative splicing event with complete exons of the canonical transcript.
Bottom figure shows exons aligned with protein domains of the gene.
Panel A illustrates an outlier event in TIF1 subgroup. Panel B illustrates an event only seen in inclusion body myositis (IBM)
To cite this abstract in AMA style:
Najjar R, Pinal-Fernandez I, Mammen A, Mustelin T. Alternative Splicing and Expected Protein Changes in Muscle Biopsies from Different Types of Idiopathic Inflammatory Myopathies [abstract]. Arthritis Rheumatol. 2023; 75 (suppl 9). https://acrabstracts.org/abstract/alternative-splicing-and-expected-protein-changes-in-muscle-biopsies-from-different-types-of-idiopathic-inflammatory-myopathies/. Accessed .« Back to ACR Convergence 2023
ACR Meeting Abstracts - https://acrabstracts.org/abstract/alternative-splicing-and-expected-protein-changes-in-muscle-biopsies-from-different-types-of-idiopathic-inflammatory-myopathies/