Session Information
Session Type: Abstract Session
Session Time: 10:00AM-10:50AM
Background/Purpose: Dermatomyositis (DM) is an autoimmune myopathy associated with marked microvascular dysfunction and high morbidity and mortality. The gut microbiome has been implicated in the pathogenesis of both vascular and autoimmune disease, however, no studies to date have evaluated whether the gut microbiome is altered in patients with dermatomyositis.
Methods: Fecal samples from DM patients and healthy controls underwent 16S rRNA gene sequencing by Illumina MiSeq. Amplicon sequence variants were identified using DADA2 and the SILVA 132 database. Microbial composition was compared between DM and control groups (beta diversity analysis) using the robust Aitchison distance metric and permutational multivariate analysis of variance controlled for significant differences between groups such as antibiotic use. Alpha diversity was measured by the Shannon Index (reflecting species richness and evenness in each sample) and was compared between groups using analysis of variance. Microbiome diversity and composition were also examined in association with several clinical variables including myositis-specific autoantibodies (MSA). Differentially abundant microbial taxa associated with clinical characteristics were identified using DESeq2.
Results: A total of 36 DM patients and 26 healthy controls (Table 1) were analyzed. Compared with controls, DM patients exhibited a trend towards lower microbial alpha diversity based on the Shannon index (p= 0.08). Higher disease damage (measured by physician global visual analogue scales) in DM patients was significantly correlated with lower microbial diversity (Figure 1-d). When DM patients were analyzed by MSA subgroups (ILD-associated MSA: antisynthetase ab,anti-MDA5 ab, n=12; Cancer-associated: anti-NXP2 ab, anti-TIF1gamma ab, n=13) and compared with controls, there were significant differences in their microbial composition (Figure 2-a). This was also associated with significantly reduced Shannon index in the ILD-associated MSA group compared to controls (Figure 2-b). Differential abundance testing demonstrated that several amplicon sequence variants within the Firmicutes, Actinobacteriaand Bacteroidetes phyla were found in greater abundance in ILD-associated and cancer-associated MSA patients compared to controls (Figure 2-d).
Conclusion: DM patients with ILD-associated MSAs have an altered gut microbiome including significantly decreased microbial diversity. Further studies are warranted to determine whether these abnormalities contribute to disease pathogenesis including microvascular damage.
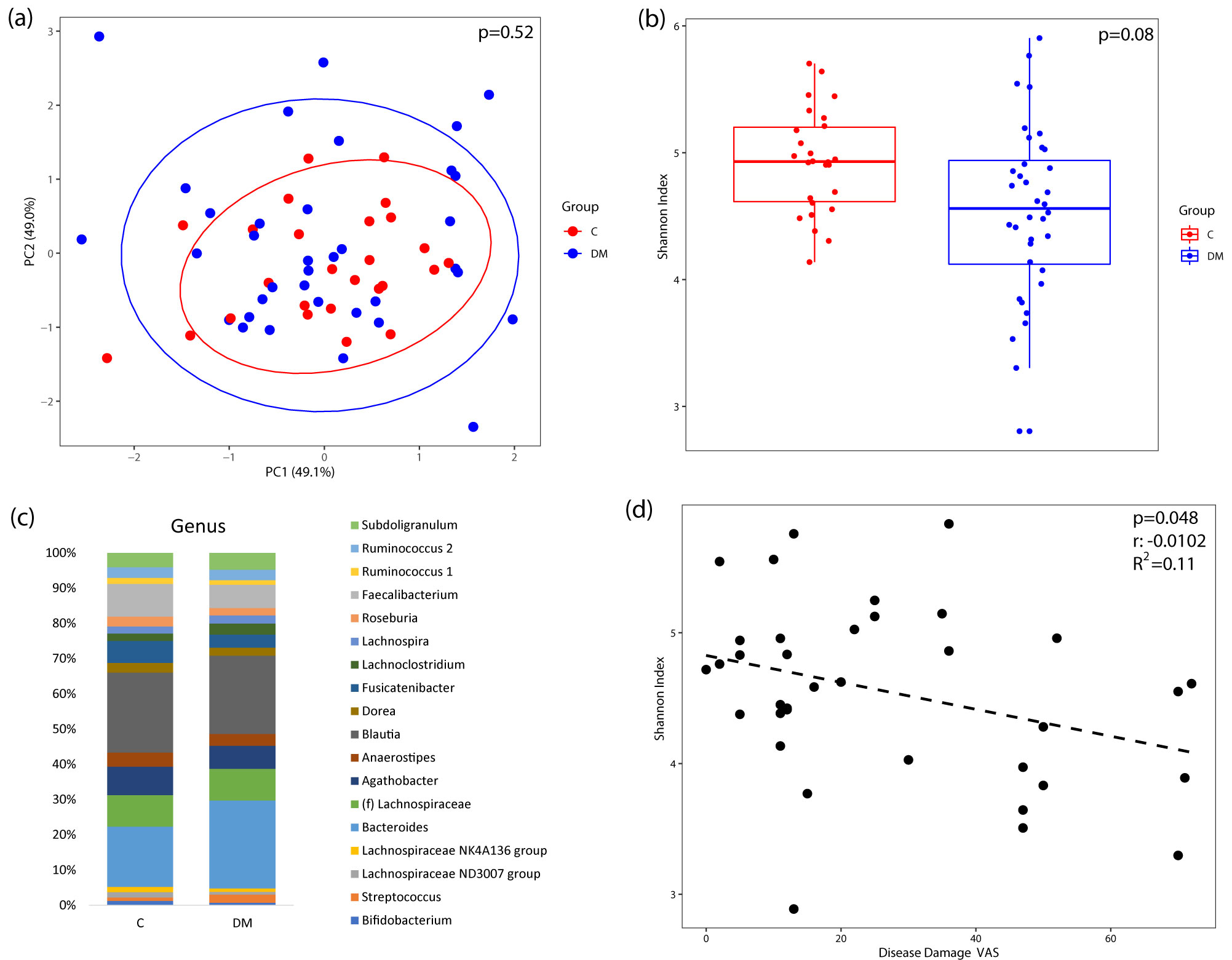 Figure 1. Microbiome diversity in DM patients (n=36) compared to healthy controls (n=26). (a) Comparing the overall microbial composition in DM and healthy control samples by principal components analysis plot using a robust Aitchison distance matrix. Each dot represents a sample from a DM patient (blue) or a healthy control (red). Ellipse represents the 95% confidence interval for each group. (b) Species evenness and richness in DM vs controls by Shannon index adjusted for significant differences in antibiotic use. p value = 0.08 (c) Taxa summary plots between DM and control with only those genera with a relative abundance of ≥ 1%. No genus was differentially abundant in DM compared to controls. (d) Higher disease damage score by visual analog scale in DM patients was significantly correlated with lower evenness and diversity of microbial species.
Figure 1. Microbiome diversity in DM patients (n=36) compared to healthy controls (n=26). (a) Comparing the overall microbial composition in DM and healthy control samples by principal components analysis plot using a robust Aitchison distance matrix. Each dot represents a sample from a DM patient (blue) or a healthy control (red). Ellipse represents the 95% confidence interval for each group. (b) Species evenness and richness in DM vs controls by Shannon index adjusted for significant differences in antibiotic use. p value = 0.08 (c) Taxa summary plots between DM and control with only those genera with a relative abundance of ≥ 1%. No genus was differentially abundant in DM compared to controls. (d) Higher disease damage score by visual analog scale in DM patients was significantly correlated with lower evenness and diversity of microbial species.
 Figure 2. Microbiome diversity in DM patients by MSA subgroups (ILD associated MSA n=12, Cancer associated MSA n=13) compared to healthy controls (n=26). (a) Significant differences in the overall microbial composition between MSA/MAA subgroups of DM and controls. Each dot represents a sample from a DM patient with ILD associated MSA (green), DM patient with cancer associated MSA (red) or healthy control (blue). Ellipse represents the 95% confidence interval for each group. (b) Decreased species evenness and richness in MSA/MAA subgroups of DM compared to controls. (c) Taxonomic summary plots by MSA subgroups, only showing those genera with a relative abundance ≥ 1%. Differential abundance testing at the amplicon sequence variant level between (d) cancer associated MSA patients vs control, (e) ILD associated MSA patients vs control, and (f) ILD associated MSA patients vs Cancer associated MSA patients.
Figure 2. Microbiome diversity in DM patients by MSA subgroups (ILD associated MSA n=12, Cancer associated MSA n=13) compared to healthy controls (n=26). (a) Significant differences in the overall microbial composition between MSA/MAA subgroups of DM and controls. Each dot represents a sample from a DM patient with ILD associated MSA (green), DM patient with cancer associated MSA (red) or healthy control (blue). Ellipse represents the 95% confidence interval for each group. (b) Decreased species evenness and richness in MSA/MAA subgroups of DM compared to controls. (c) Taxonomic summary plots by MSA subgroups, only showing those genera with a relative abundance ≥ 1%. Differential abundance testing at the amplicon sequence variant level between (d) cancer associated MSA patients vs control, (e) ILD associated MSA patients vs control, and (f) ILD associated MSA patients vs Cancer associated MSA patients.
To cite this abstract in AMA style:
Bae S, Dong T, Lagishetty V, Katzka W, Jacobs J, Charles-Schoeman C. Altered Gut Microbiome in Dermatomyositis [abstract]. Arthritis Rheumatol. 2020; 72 (suppl 10). https://acrabstracts.org/abstract/altered-gut-microbiome-in-dermatomyositis/. Accessed .« Back to ACR Convergence 2020
ACR Meeting Abstracts - https://acrabstracts.org/abstract/altered-gut-microbiome-in-dermatomyositis/

