Session Information
Date: Tuesday, October 28, 2025
Title: Abstracts: Osteoarthritis & Joint Biology – Basic Science (1734–1739)
Session Type: Abstract Session
Session Time: 11:15AM-11:30AM
Background/Purpose: In arthritic synovium, emerging clinical evidence suggests crosstalk between synovial cells and dorsal root ganglion (DRG) neurons drives nerve sprouting and pain sensitization [Bai+ 2024, Nanus+ 2021]. Similarly, preclinical post-traumatic osteoarthritis (PTOA) models demonstrate progressive knee hyperalgesia and rapid nociceptor sprouting post-injury [Bergman+ 2023, Obeidat+2024]. To elucidate the synovium-nerve signaling underpinning injury-induced nociceptor sprouting, we used bulk RNA sequencing to characterize the acute response of paired synovium and DRG to ACL rupture (ACLR).
Methods: Under IACUC approval, 12-week-old male C57Bl/6 mice underwent ACLR or sham procedure (n=6). At 1, 3, or 7 days post-injury, total RNA was isolated from ipsilateral synovium and L3-L5 DRGs for bulk RNA sequencing. Quality control confirmed high RNA and sequencing quality and high DRG neuronal enrichment (Fig 1A,B). Data were analyzed for differentially-expressed genes (DEGs) via weighted Limma-Voom, and pathway enrichment via gene set enrichment analysis (GSEA). A published DRG interactome database [Wangzhou+ 2021] was utilized to identify putative interactions between ACLR-upregulated synovial ligands and DRG-neuron receptors. Reference single cell RNA sequencing (scRNAseq) data of murine synovium and DRG was used to assess cell-specific ligand/receptor expression.
Results: The synovium exhibited a marked, gradually-evolving response to ACLR (Fig 1C,D). Most injury-induced DEGs ( >60%) were conserved across timepoints (Fig 1E), with >10% shared between adjacent timepoints (Fig 1E). Angio- and neurogenic synovial signaling was detectable at 1d and progressive out to 7d (Fig 1D,E). Inflammatory signaling was most enriched at 1d but sustained throughout (Fig 1D). Neuroinflammatory leading edge genes suggested multifaceted neuro-immune signaling (e.g. complement (C5ar1), toll-like receptor (Tlr7), and cytokine (Tnfrsf1b) pathways, Fig 1E). Progressive fibroblast-associated matrix changes denoted initiation of progressive fibrosis, hyperplasia, and ossification known to present at later timepoints in this model (Fig 1D). DRG response to ACLR was comparatively subtler (Fig 2A-C), but GSEA revealed significant enrichment of several neuron sprouting pathways (Fig 2D,E) across timepoints, with different facets preferentially enriched at 1, 3, and 7 days post-ACLR (Fig 2D), suggesting a rapid, evolving post-injury response. Interactome analysis (Fig 3A) implicated pathways including NGF, semaphorin-plexin, FGF and integrin signaling as putative mediators of time-dependent (Fig 3C), injury-induced synovium-nerve crosstalk. scRNAseq demonstrated synovial ligand secretion primarily by fibroblasts, Schwann cells, and pericytes (Fig 3B), with receptor expression by various neuronal subsets (Fig 3D).
Conclusion: These data reveal a rapid, injury-induced neuroinflammatory synovial response concomitant with transcriptional programs indicative of neuronal plasticity in the DRG. Putative synovium-DRG interactome analysis implicated pathways such as NGF and sema-plex as potential axes of synovium-DRG crosstalk underlying injury-induced nociceptor sprouting.
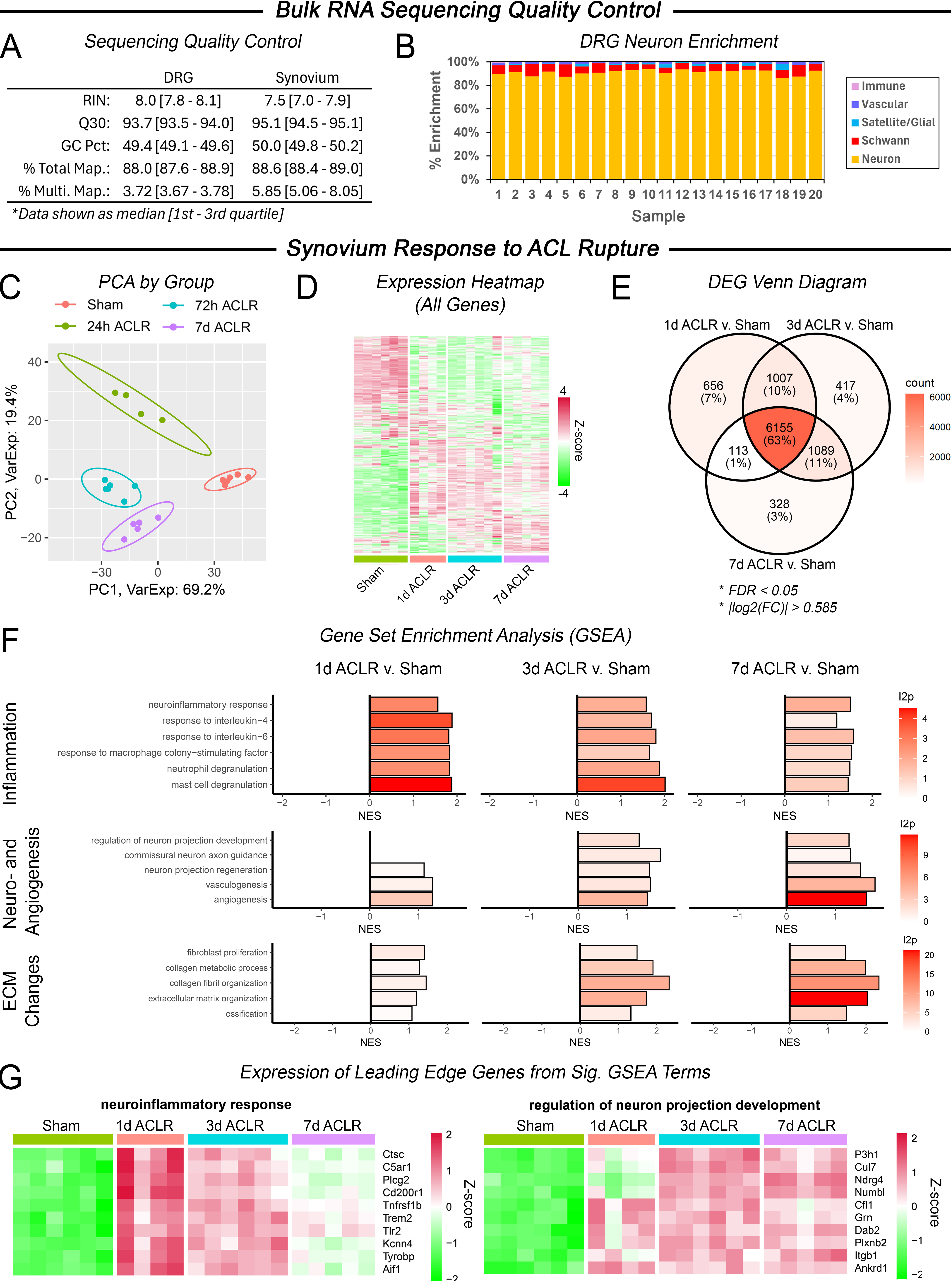 Figure 1. (A) Bulk RNA sequencing quality control metrics for DRG and synovium. (B) DRG neuron enrichment, calculated via bulk seq. deconvolution. Synovium response to ACL rupture: (C) PCA plot. (D) Heatmap of all genes. (E) Venn diagram of DEG overlap between timepoints. (F) Significant GSEA terms related to inflammation, neuroangiogenesis, and ECM. (G) Expression of leading-edge genes from significant GSEA terms.
Figure 1. (A) Bulk RNA sequencing quality control metrics for DRG and synovium. (B) DRG neuron enrichment, calculated via bulk seq. deconvolution. Synovium response to ACL rupture: (C) PCA plot. (D) Heatmap of all genes. (E) Venn diagram of DEG overlap between timepoints. (F) Significant GSEA terms related to inflammation, neuroangiogenesis, and ECM. (G) Expression of leading-edge genes from significant GSEA terms.
.jpg) Figure 2. DRG response to ACL rupture (A) PCA plot. (B) Heatmap of expression across all genes. (C) Venn diagram of DEG overlap between timepoints. (D) Significant GSEA terms related to neuron sprouting and synapse regulation (E) Expression of leading-edge genes from significant GSEA terms.
Figure 2. DRG response to ACL rupture (A) PCA plot. (B) Heatmap of expression across all genes. (C) Venn diagram of DEG overlap between timepoints. (D) Significant GSEA terms related to neuron sprouting and synapse regulation (E) Expression of leading-edge genes from significant GSEA terms.
.jpg) Figure 3. Synovium-DRG interactome. (A) Alluvial plots showing potential interactions between significantly injury-upregulated, secreted synovial ligands and membrane-bound, DRG neuron-derived receptors. (B) Synovial ligand expression by post-injury time point and cell type, generated using a reference scRNAseq dataset of murine synovium. (C) Overlap in interactions by post-injury timepoint. (D) DRG neuron expression by neuron subset, generated using a reference scRNAseq dataset of murine DRG.
Figure 3. Synovium-DRG interactome. (A) Alluvial plots showing potential interactions between significantly injury-upregulated, secreted synovial ligands and membrane-bound, DRG neuron-derived receptors. (B) Synovial ligand expression by post-injury time point and cell type, generated using a reference scRNAseq dataset of murine synovium. (C) Overlap in interactions by post-injury timepoint. (D) DRG neuron expression by neuron subset, generated using a reference scRNAseq dataset of murine DRG.
To cite this abstract in AMA style:
Newton M, Wang L, Malfait A, Miller R, Maerz T. Acute Transcriptomic Response of Synovium and Dorsal Root Ganglia to Joint Injury [abstract]. Arthritis Rheumatol. 2025; 77 (suppl 9). https://acrabstracts.org/abstract/acute-transcriptomic-response-of-synovium-and-dorsal-root-ganglia-to-joint-injury/. Accessed .« Back to ACR Convergence 2025
ACR Meeting Abstracts - https://acrabstracts.org/abstract/acute-transcriptomic-response-of-synovium-and-dorsal-root-ganglia-to-joint-injury/
