Session Information
Date: Monday, October 27, 2025
Title: (0955–0977) Systemic Sclerosis & Related Disorders – Basic Science Poster I
Session Type: Poster Session B
Session Time: 10:30AM-12:30PM
Background/Purpose: Autoimmune conditions with skin involvement, including scleroderma (SSc), antiphospholipid syndrome (APS), systemic lupus erythematosus (SLE) and psoriasis (PSO), involve intricate interactions between immune cells and the vasculature, with endothelial cells (ECs) playing a critical role in disease pathogenesis. To elucidate the diverse functional roles of ECs in these conditions, we constructed a comprehensive single-cell atlas by integrating single-cell RNA sequencing (scRNA-seq) data from affected and healthy skin biopsies.
Methods: Skin biopsies from patients were analyzed using 10x Genomics scRNA-seq. Publicly available datasets from SSc (GSE249279), APS (GSE290946), SLE (GSE186476), PSO (GSE173706), and healthy controls (GSE249279 and GSE290946), generated using the same platform, were integrated (Figure 1A). Skin biopsies from active skin of patients were analyzed using 10x Genomics scRNA-seq. Cell identities, gene expression profiles, and pathway activities were analyzed across endothelial and stromal populations.
Results: We profiled 23,410 endothelial and stromal cells, identifying arterial, venous, capillary, and lymphatic ECs, as well as pericytes and vascular smooth muscle cells (VSMCs) from all scRNA-seq datasets. Arterial and venous ECs were significantly reduced in SSc, while VSMCs were markedly increased. APS samples showed an expansion of arterial, capillary and venous ECs, whereas SLE and PSO exhibited cellular compositions similar to healthy controls (Figure 1). Pathway enrichment analysis of EC gene expression revealed both shared and disease-specific programs (Figure 2). Across autoimmune conditions, common signatures relative to healthy controls included NF-κB signaling, leukocyte adhesion, and antigen presentation. APS-derived ECs were further enriched for antigen presentation, oxidized LDL response, leukocyte adhesion, and T cell activation pathways. SLE samples demonstrated prominent type I interferon responses, while SSc samples exhibited strong enrichment of intrinsic apoptotic pathways. No dominant pathway activation was observed in PSO.Deeper subclustering of vascular ECs defined seven endothelial subtypes, including SENA3G+ arterial ECs, RGCC+ capillary ECs, TLL1⁺ venous ECs, hypoxia-associated ECs, EndoMT, cycling ECs, and a previously undefined subset IL6⁺ venous ECs (Figure 3). IL6⁺ and EndoMT ECs were enriched in SSc and expressed pro-inflammatory, senescence-associated, and matrix remodeling genes. SLE and PSO had elevated IL6+ EC and cycling EC populations, potentially driving immune dysregulation and vascular proliferation linked to systemic organ damage. APS was marked by abundant EndoMT+ ECs, suggesting endothelial plasticity. Senescence and EndoMT scores were highest in SSc, followed by APS and SLE, and minimal in PSO.
Conclusion: This single-cell atlas defines dermal endothelial heterogeneity across autoimmune diseases. Notably, we identified a novel IL6⁺ EC subset enriched in SSc with pro-inflammatory and senescence signatures, and revealed distinct EC programs across diseases. These findings highlight EC subtype dynamics as potential contributors to immune dysregulation and fibrotic remodeling.
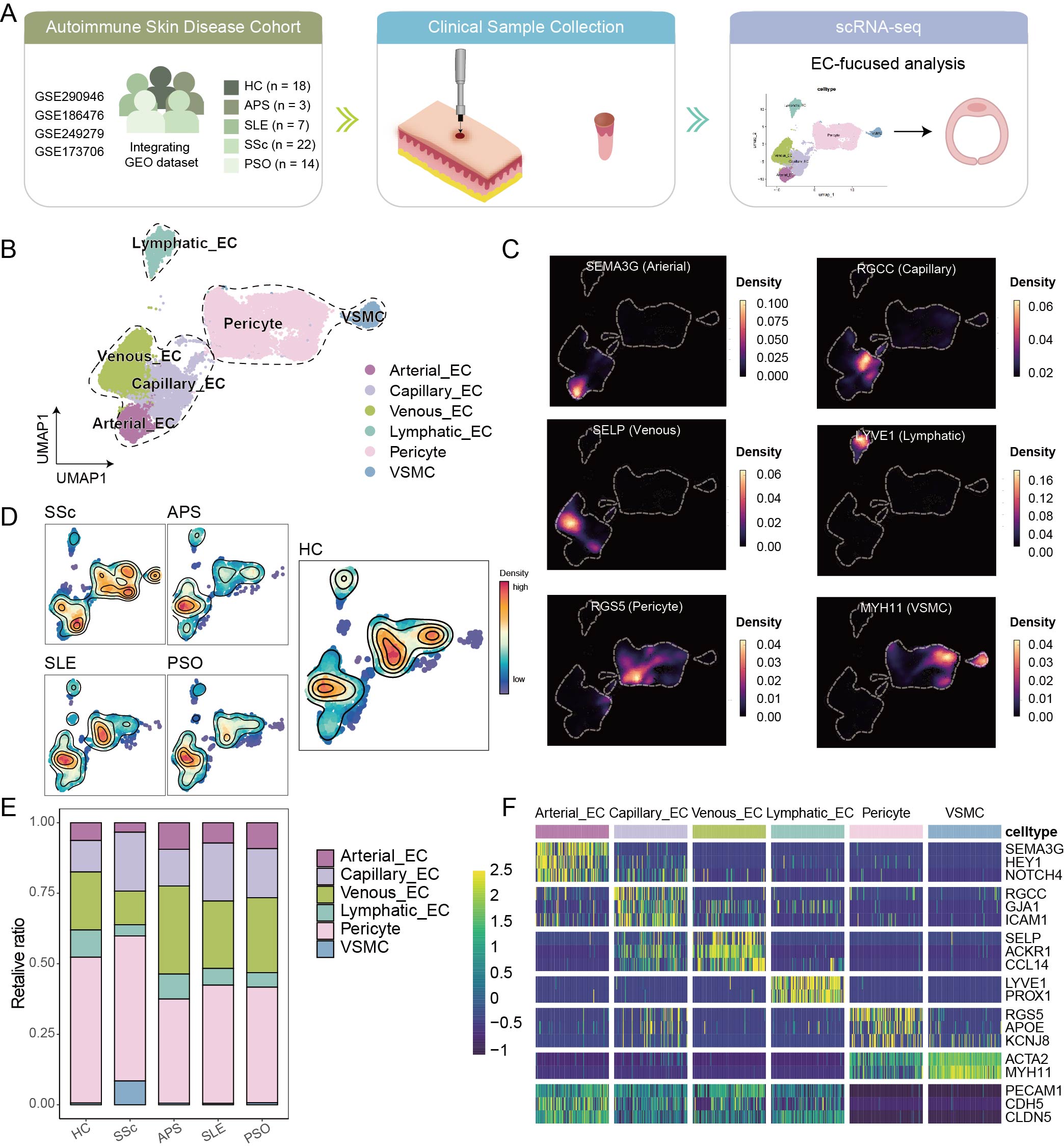 Figure 1. Cellular composition and endothelial subtypes across autoimmune skin diseases. (A) Schematic overview of the study design integrating single-cell RNA sequencing data from autoimmune skin diseases including APS, SLE, PSO, and SSc, along with healthy controls. (B) UMAP visualization of 23,410 endothelial and stromal cells from skin biopsies, annotated into arterial, venous, capillary, and lymphatic ECs, pericytes, and vascular smooth muscle cells (VSMCs). (C) Expression density of canonical markers confirming subtype identities, such as SEMA3G (arterial EC), RGCC (capillary EC), SELP (venous EC), LYVE1 (lymphatic EC), MYH11 (VSMC), and RGS5 (pericyte). (D, E) Relative abundance of vascular and stromal subtypes across disease groups. (F) Heatmap showing the averaged expression of key markers across identified cell types.
Figure 1. Cellular composition and endothelial subtypes across autoimmune skin diseases. (A) Schematic overview of the study design integrating single-cell RNA sequencing data from autoimmune skin diseases including APS, SLE, PSO, and SSc, along with healthy controls. (B) UMAP visualization of 23,410 endothelial and stromal cells from skin biopsies, annotated into arterial, venous, capillary, and lymphatic ECs, pericytes, and vascular smooth muscle cells (VSMCs). (C) Expression density of canonical markers confirming subtype identities, such as SEMA3G (arterial EC), RGCC (capillary EC), SELP (venous EC), LYVE1 (lymphatic EC), MYH11 (VSMC), and RGS5 (pericyte). (D, E) Relative abundance of vascular and stromal subtypes across disease groups. (F) Heatmap showing the averaged expression of key markers across identified cell types.
.jpg) Figure 2. Transcriptional and pathway-level profiling of endothelial and stromal cells across autoimmune skin diseases. (A) Multidimensional scaling (MDS) plots of endothelial and stromal cell subsets showing global transcriptomic distribution across APS, SLE, PSO, and SSc. (B) Volcano plots showing differentially expressed genes in vascular ECs, pericytes, VSMCs, and lymphatic ECs across disease groups. (C) Radar plots depicting pathway enrichment scores of arterial, venous, and capillary ECs across eight biological processes.
Figure 2. Transcriptional and pathway-level profiling of endothelial and stromal cells across autoimmune skin diseases. (A) Multidimensional scaling (MDS) plots of endothelial and stromal cell subsets showing global transcriptomic distribution across APS, SLE, PSO, and SSc. (B) Volcano plots showing differentially expressed genes in vascular ECs, pericytes, VSMCs, and lymphatic ECs across disease groups. (C) Radar plots depicting pathway enrichment scores of arterial, venous, and capillary ECs across eight biological processes.
.jpg) Figure 3. Endothelial subcluster identification and their distribution across autoimmune skin diseases. (A) UMAP projection of endothelial cells showing seven transcriptionally distinct subclusters: SEMA3G⁺ ECs, RGCC⁺ ECs, TLL1⁺ ECs, IL6⁺ ECs, hypoxia-associated ECs, EndoMT ECs, and cycling ECs. (B) Spatial expression of canonical endothelial and subcluster marker genes, including PECAM1, SEMA3G, RGCC, and ACKR1. (C) Dot plot displaying average expression (color) and proportion of cells (size) for representative marker genes across endothelial subclusters. (D) Disease preference (HC, SSc, APS, SLE, PSO) of each cluster estimated by the STARTRAC-dist index. Ro/e denotes the ratio of observed to expected cell number. (E) Distribution of senescence scores across endothelial subclusters and disease conditions. (F) Distribution of EndoMT scores across endothelial subclusters and disease groups.
Figure 3. Endothelial subcluster identification and their distribution across autoimmune skin diseases. (A) UMAP projection of endothelial cells showing seven transcriptionally distinct subclusters: SEMA3G⁺ ECs, RGCC⁺ ECs, TLL1⁺ ECs, IL6⁺ ECs, hypoxia-associated ECs, EndoMT ECs, and cycling ECs. (B) Spatial expression of canonical endothelial and subcluster marker genes, including PECAM1, SEMA3G, RGCC, and ACKR1. (C) Dot plot displaying average expression (color) and proportion of cells (size) for representative marker genes across endothelial subclusters. (D) Disease preference (HC, SSc, APS, SLE, PSO) of each cluster estimated by the STARTRAC-dist index. Ro/e denotes the ratio of observed to expected cell number. (E) Distribution of senescence scores across endothelial subclusters and disease conditions. (F) Distribution of EndoMT scores across endothelial subclusters and disease groups.
To cite this abstract in AMA style:
Pan H, Qian J, Yu S, Yang Z, Knight J, Tsou E, Shi H. A Single-Cell Atlas Reveals Dermal Endothelial Heterogeneity and Disease-Specific Pathways in Autoimmune Disorders [abstract]. Arthritis Rheumatol. 2025; 77 (suppl 9). https://acrabstracts.org/abstract/a-single-cell-atlas-reveals-dermal-endothelial-heterogeneity-and-disease-specific-pathways-in-autoimmune-disorders/. Accessed .« Back to ACR Convergence 2025
ACR Meeting Abstracts - https://acrabstracts.org/abstract/a-single-cell-atlas-reveals-dermal-endothelial-heterogeneity-and-disease-specific-pathways-in-autoimmune-disorders/
