Session Information
Session Type: Poster Session C
Session Time: 1:00PM-3:00PM
Background/Purpose: Knee osteoarthritis progression is heterogeneous in the population, being the most aggresive form the rapid progressive knee osteoarthritis (RPKOA). These patients suffer a worsening of the joint in a short window of time. Previous results showed that mitochondrial DNA (mtDNA) variation modify the incidence and progression of OA, but their influence on the RPKOA is unknown. Our objectives for the present work are:
- To find new mtDNA variants related to RPKOA to develop a predictive model
- To find out the functional significance of these variants
Methods: In the discovery phase we used the Osteoarthritis Initiative (OAI) and in the replication phase we pooled individuals from the Cohort Hip and Cohort Knee (CHECK) and the Prospective Cohort of Osteoarthritis from A Coruña (PROCOAC) cohorts. Patients were classified as follows:
- Rapid progressors (N Discovery=268, N Replication=107), subjects with baseline KL grade 0-1 and increase up to KL≥3 during 48-month follow-up; or baseline KL grade 2 and increase up to KL grade 4 during the follow-up
- Non-rapid progressors (N Discovery=827, N Replication=592), subjects with the same baseline characteristics as rapid progressors, but with a slower or no evolution over time
mtDNA in the OAI was screened by in-depth sequencing. Resulting variant was analyzed in the replication phase by Sanger sequencing. Statistical approaches adjusting by confounding variables followed by a meta-analysis were applied. We developed a functional study using two types of transmitochondrial cybrids belonging to mtDNA haplogroup H, with and without the mitochondrial variant of interest.
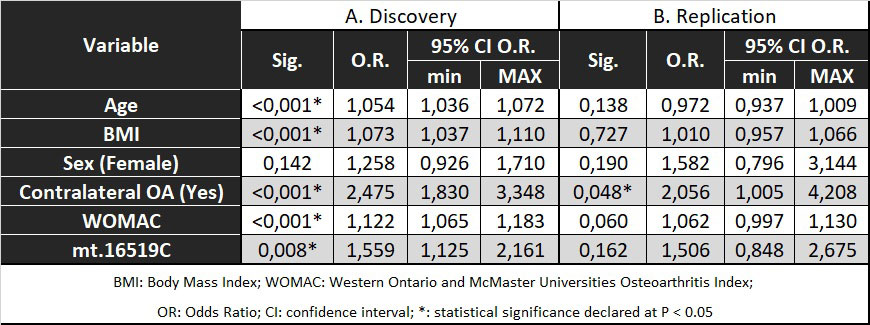
Results: The in-depth sequencing of mtDNA revealed that mtDNA D-loop variant m.16519C showed a significantly higher frequency in the rapid-progressors group (OR=1,558;95%CI=1,148-2,113;nominal p-value=0,004; B-H corrected pvalue=0,044). These results were similar to those obtained in the replication cohort (OR=1,647;95%CI=1,013-2,678; p=0,043).
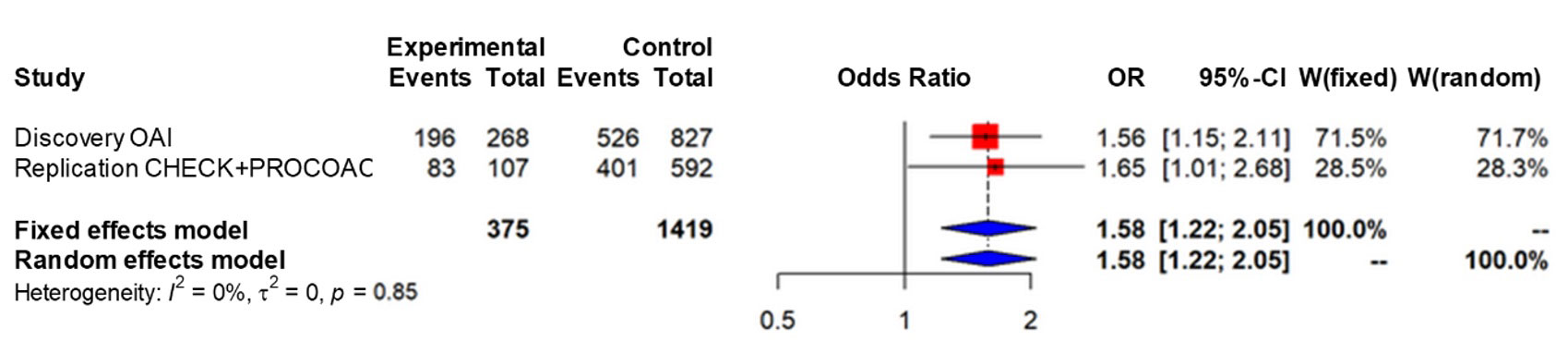
The preditive models of the OAI revealed that, regardless of the different clinical variables, the variant m.16519C was significantly associated with RPKOA (OR=1,559;95%CI=1,125-2,161; p=0,008). By contrast, in the replication phase the trend was similar but bordered on the statistical significance (OR=1,506;95%CI=0,848-2,675;p=0,162) (Table 1). The subsequent meta-analysis confirmed the significant association between this mtDNA variant and RPKOA (OR=1,58;95% CI=1,22-2,05;p=0,0027) (Figure 1).
In contrast with wildtype cybrids, those harboring the mtDNA variant showed lower expression mRNA levels of PGC-1α (p=0,0024) and FIS1 (p=0,0519), as well as a lower average cell survival under oxidative stress conditions (23,42±8,88 vs 115,7±64,1;p=0,0043). In addition, cybrind harbouring this variant alsho showed a increased mitochondrial reactive oxygen species (ROS) production (4,21±1,29 vs 2,21±1,34;p=0,014).
Conclusion: D-loop variant m.16519C increases the risk of developing a rapid progression of knee OA. Among the effects caused by this variant both mitochondrial biogenesis and fission/fusion processes, as well as cell survival under oxidative stress conditions and mitochondrial ROS production, seem to be affected.
To cite this abstract in AMA style:
Durán-Sotuela A, Fernández-Moreno M, Hermida-Gómez T, Relaño S, Suárez-Ulloa V, Balboa-Barreiro v, Oreiro N, Vázquez-García J, Blanco F, Rego-Pérez I. A Novel Mitochondrial Variant as a Predictor for the Rapid Progressive Knee Osteoarthritis Phenotype: Development of a Predictive Model and Functional Studies [abstract]. Arthritis Rheumatol. 2022; 74 (suppl 9). https://acrabstracts.org/abstract/a-novel-mitochondrial-variant-as-a-predictor-for-the-rapid-progressive-knee-osteoarthritis-phenotype-development-of-a-predictive-model-and-functional-studies/. Accessed .« Back to ACR Convergence 2022
ACR Meeting Abstracts - https://acrabstracts.org/abstract/a-novel-mitochondrial-variant-as-a-predictor-for-the-rapid-progressive-knee-osteoarthritis-phenotype-development-of-a-predictive-model-and-functional-studies/