Session Information
Date: Monday, October 27, 2025
Title: Abstracts: B Cell Biology & Targets in Autoimmune & Inflammatory Disease (1691–1697)
Session Type: Abstract Session
Session Time: 4:00PM-4:15PM
Background/Purpose: Most genetic variants that confer risk of complex autoimmune diseases affect gene regulation in specific cell types. Their target genes and focus cell types are often unknown, partially because some effects are hidden in untested cell states. B cells play important roles in autoimmune diseases, including systemic lupus erythematosus (SLE), rheumatoid arthritis, and multiple sclerosis. However, despite this established importance, B cell activation states are underrepresented in functional genomics studies.
Methods: In this study, we obtained B cells from 26 healthy female donors. We stimulated B cells in vitro with six activation conditions targeting key pathways: the B cell receptor (BCR), Toll-like receptor 7 (TLR7), TLR9, CD40, and a cocktail that promotes differentiation into double negative 2 (DN2) IgD– CD27– CD11c+ CD21– B cells, a likely pathogenic B cell subset for autoimmunity (Fig. 1). We profiled up to 24 human B cell activation states and up to 5 control conditions using RNA-seq, single-cell RNA-seq coupled with surface protein markers (CITE-seq), and ATAC-seq.
Results: We provide an in-depth characterization of how autoimmune disease-associated genes respond to stimuli and group into modules with distinct functions (Fig. 2a-c). Using high-depth RNA-seq data, we find pervasive splicing effects during B cell activation. Using single-cell data, we describe stimulus-dependent B cell fates (Fig. 2d-g). Chromatin data reveal the transcription factors likely involved in stimulus-dependent B cell activation. These open chromatin regions capture a significant proportion of the genetic risk of multiple autoimmune diseases and elucidate autoimmune disease genetic risk variants with previously unknown function. To validate the utility of our findings, we performed fine-mapping for an SLE risk locus and identified a candidate causal variant (Fig. 3a) that overlaps an open chromatin region specific for B cells activated with the DN2-cocktail (Fig. 3b). This intergenic open chromatin peak is 15kb away from TNFSF4, which codes for OX40L and mediates interaction with activated T cells. TNFSF4 gets upregulated with DN2-cocktail activation at 48 and 72 hours (Fig. 3c). We used CRISPR inactivation in a B cell line to silence the putative enhancer where the SLE risk variant is located, which demonstrated that its target gene is TNFSF4 (Fig. 3d). We have made our multiomics resource available via an interactive browser that can be used to query the dynamics of gene regulation and B cell differentiation during activation by different stimuli, enhancing further investigation of B cells and their role in autoimmune diseases.
Conclusion: Our B cell activation multi-omics resource reveals how B cells are transcriptionally and epigenetically wired to respond to distinct stimuli, highlighting pathway-specific activation programs. It also offers mechanistic insights into the genetic basis of autoimmune diseases, including functional validation of a lupus-associated risk locus.
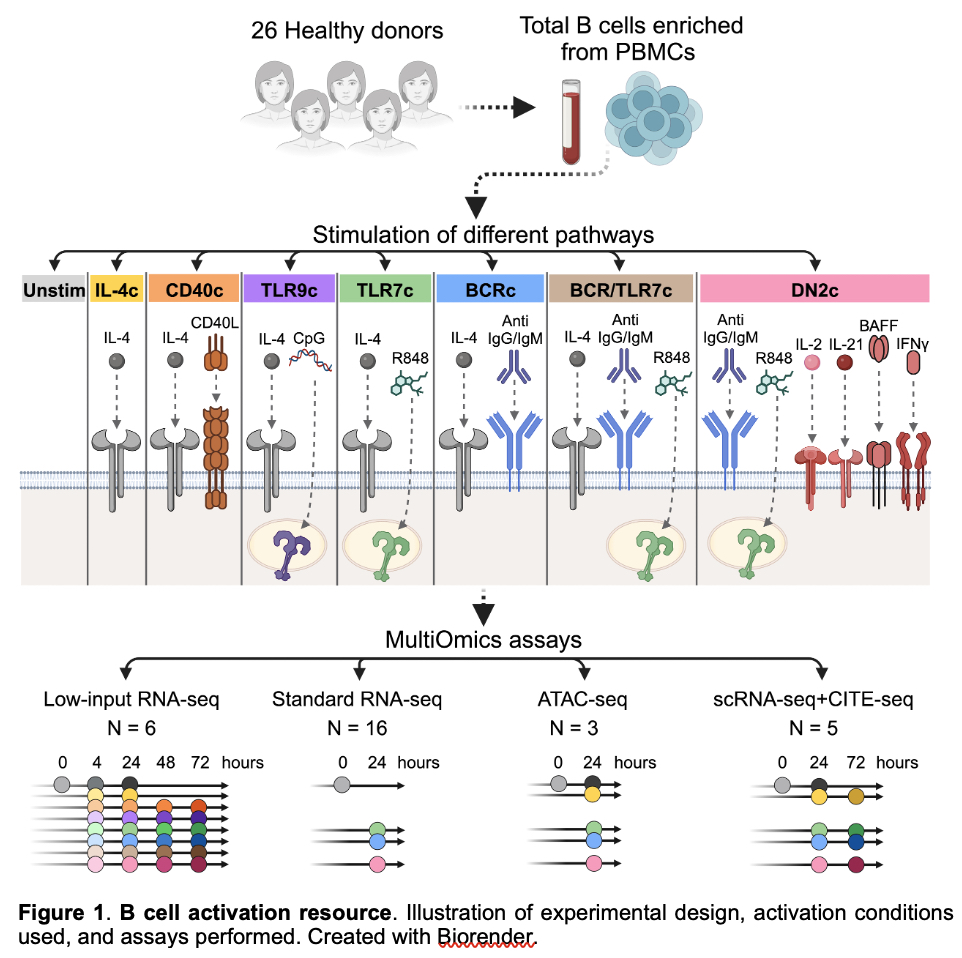 Figure 1. B cell activation resource. Illustration of experimental design, activation conditions used, and assays performed. Created with Biorender.
Figure 1. B cell activation resource. Illustration of experimental design, activation conditions used, and assays performed. Created with Biorender.
.jpg) Figure 2. Dynamic gene expression in activated B cells over time and pathway-dependent B cell fates. a, Principal component analysis (PCA) with the top 2,000 most variable genes shows separation of conditions and time points. Inset: PCA plot highlighting conditions that include BCR stimulation. b, Expression levels of selected B cell activation genes. c, Modules of gene expression identified by Weighted Gene Correlation Network Analysis (WGCNA). X-axis: time in hours. On the right, enrichment of Gene Ontology (GO) biological processes for modules. d, Uniform Manifold Approximation and Projection (UMAP) based on gene expression after Harmony correction. e, Proportion of B cell subsets defined by expression of marker protein/genes (genes in italic) for each condition and time point (colors as in (d)). f, UMAP as in (d) colored by protein expression levels of key markers. g, UMAP as in (d) colored by gene expression levels of autoimmunity-associated genes.
Figure 2. Dynamic gene expression in activated B cells over time and pathway-dependent B cell fates. a, Principal component analysis (PCA) with the top 2,000 most variable genes shows separation of conditions and time points. Inset: PCA plot highlighting conditions that include BCR stimulation. b, Expression levels of selected B cell activation genes. c, Modules of gene expression identified by Weighted Gene Correlation Network Analysis (WGCNA). X-axis: time in hours. On the right, enrichment of Gene Ontology (GO) biological processes for modules. d, Uniform Manifold Approximation and Projection (UMAP) based on gene expression after Harmony correction. e, Proportion of B cell subsets defined by expression of marker protein/genes (genes in italic) for each condition and time point (colors as in (d)). f, UMAP as in (d) colored by protein expression levels of key markers. g, UMAP as in (d) colored by gene expression levels of autoimmunity-associated genes.
.jpg) Figure 3. TNFSF4/OX40L locus. (a) GWAS data from Langefeld et al.40 rs2205960 is the most likely causal variant. PIP: posterior inclusion probability from SuSiE.67 (b) Chromatin peaks at the genomic region in (a). (c) TNFSF4 gene expression in the same conditions shown in (b). TPM: transcripts per million. (d) CRISPRi shows that inhibiting the region containing the putative causal variant leads to TNFSF4 downregulation.
Figure 3. TNFSF4/OX40L locus. (a) GWAS data from Langefeld et al.40 rs2205960 is the most likely causal variant. PIP: posterior inclusion probability from SuSiE.67 (b) Chromatin peaks at the genomic region in (a). (c) TNFSF4 gene expression in the same conditions shown in (b). TPM: transcripts per million. (d) CRISPRi shows that inhibiting the region containing the putative causal variant leads to TNFSF4 downregulation.
To cite this abstract in AMA style:
Aguiar V, Franco M, Abdel Aziz N, Fernandez-Salinas D, Chinas M, Colantuoni M, Xiao Q, Hackert N, Liao Y, Cervantes-Diaz R, Todd M, Wauford B, Wactor A, Prahalad V, Laza-Briviesca R, Darbousset R, Wang Q, Jenks S, Cashman K, Zumaquero E, Zhu Z, Case J, Cejas P, Munoz-Gomez M, Ainsworth H, Marion M, Benamar M, Lee P, Henderson L, Chang M, Wei K, Long H, Langefeld C, Gewurz B, Sanz I, Sparks J, Meidan E, Nigrovic P, Gutierrez-Arcelus M. A multi-omics resource of B cell activation reveals genetic mechanisms for autoimmune diseases [abstract]. Arthritis Rheumatol. 2025; 77 (suppl 9). https://acrabstracts.org/abstract/a-multi-omics-resource-of-b-cell-activation-reveals-genetic-mechanisms-for-autoimmune-diseases/. Accessed .« Back to ACR Convergence 2025
ACR Meeting Abstracts - https://acrabstracts.org/abstract/a-multi-omics-resource-of-b-cell-activation-reveals-genetic-mechanisms-for-autoimmune-diseases/
