Session Information
Date: Monday, November 14, 2022
Title: Plenary III
Session Type: Plenary Session
Session Time: 11:30AM-1:00PM
Background/Purpose: Genome-wide association studies (GWAS) in gout have been relatively small (≤13,179 people with gout) and have provided little insight into the progression from hyperuricemia to gout. We present a gout GWAS that includes 120,282 people with gout.
Methods: Participants were from 13 cohorts and four ancestral groups (African 3,052 gout/77,891 non-gout; East Asian 10,729/82,807; European 100,661/2,106,003; Latina 5,840/235,847). Ancestry-specific GWAS summary statistics were meta-analysed to form the trans-ancestral GWAS. Candidate causal genes were identified by co-localization with expression quantitative trait loci (eQTL) using the Gene and Tissue Expression dataset and/or having a candidate missense causal variant and/or by MAGMA gene set analysis. IL-1β response QTL were identified using the 500FG cohort. Pathway analysis was done using KEGG and drug repositioning analysis was done using Genome for Repositioning Drugs.
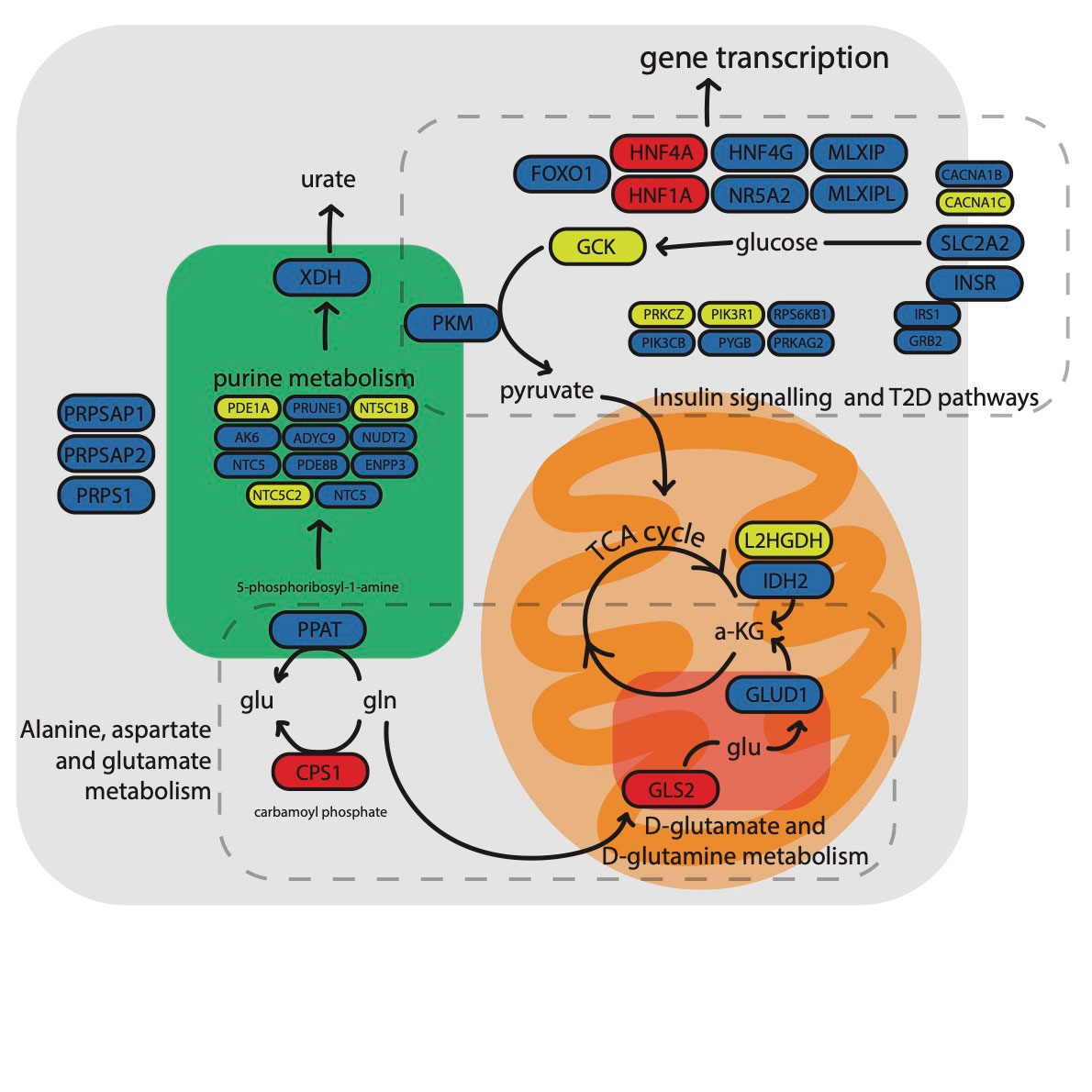
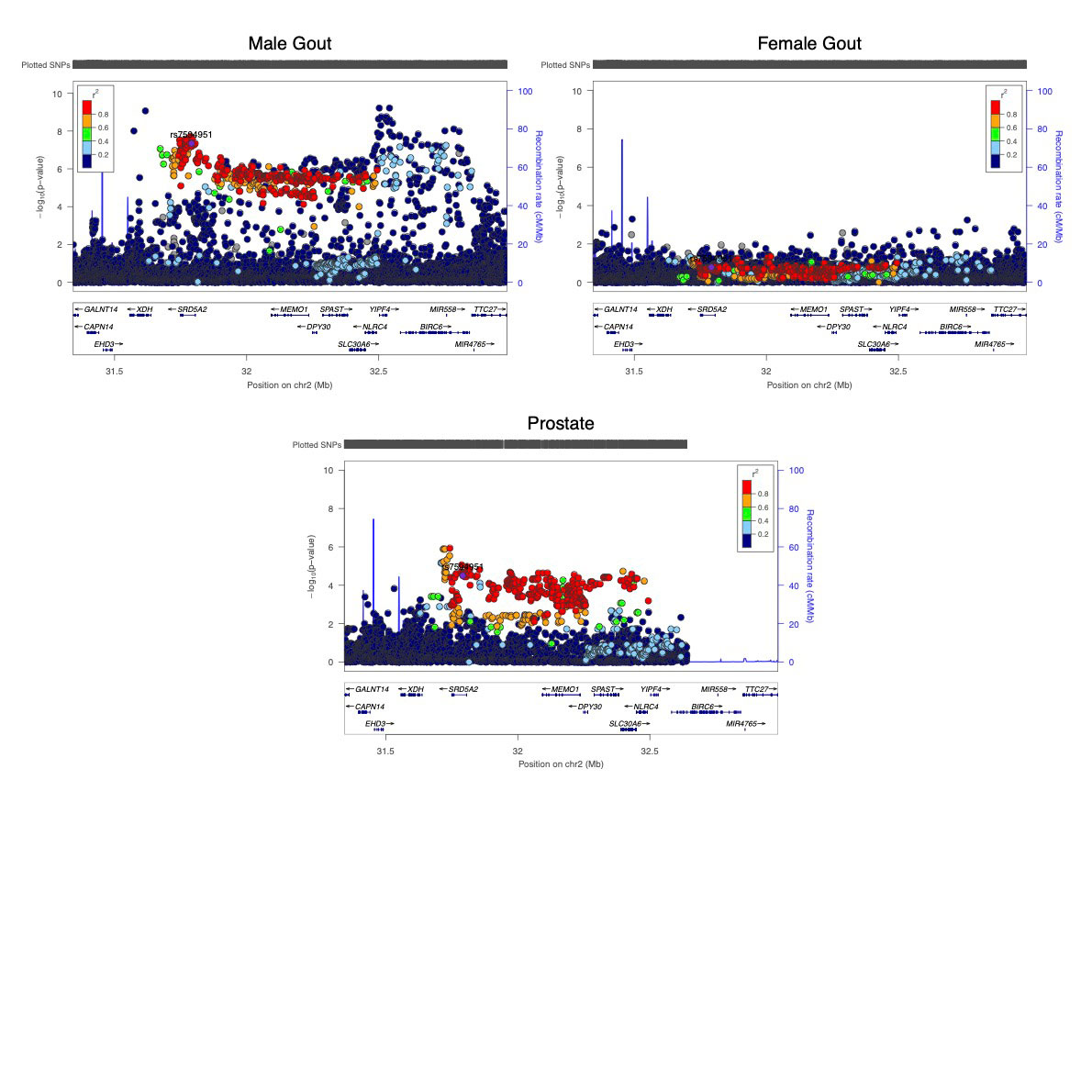
Results: There were 339 gout-associated loci encompassing 515 independently-associated variants. 123 loci did not overlap with any previously reported in urate or gout. Co-localization of GWAS and eQTL signals identified 1,657 cis- and 267 trans-eQTLs for 252 of the 339 loci. Some co-localized eQTL genes regulate NLRP3 inflammasome activation and activity (e.g. TMEM176B, SCAP, INSIG2 SREBF-AS1, FADS2, NLRC3). Co-localized eQTLs also included TET2, EZH2, IDH2, and RUNX1, genes mutated in Clonal Hematopoiesis of Indeterminate Potential (CHIP). Enriched molecular pathways using candidate genes identified purine metabolism, amino acid metabolism and insulin signalling pathways (Figure 1). Genetic variants present in the 500FG dataset and not previously associated with urate (n=93, possibly involved in inflammation in gout) had amplification of signal that associated with IL-1b response to monosodium urate (MSU) crystal and LPS co-stimulation in peripheral blood mononuclear cells (Figure 2). However those associated with urate (n=317) did not exhibit amplified signal (Figure 2). The gout GWAS signal at the top-associated variant of the 93 variants not associated with urate (rs9973741) co-localized with genetic control of production of IL-1b upon stimulation (the gout risk allele associated with increased IL-1β level), and also co-localized with genetic control (eQTL) of IL1RN and IL-38 expression, implicating these genes as causal at this locus. An association signal at Xanthine Dehydrogenase was restricted to males and co-localized with XDH expression (eQTL) only in the prostate (Figure 3). Gout-associated loci represented drug repurposing possibilities from neoplasm, blood biochemistry, and metabolic disorder treatment categories.
Conclusion: We implicate multiple new causal genes and pathways in gout. In addition to genes that regulate urate, NLRP3 inflammasome activity and autophagy, this work has identified the CHIP pathway, that includes epigenome modification proteins, in gout pathogenesis. Other pathways identified involve insulin signalling and purine and glutamate metabolism. Uniquely regulated production of urate by the prostate potentially represents a novel risk factor for gout in men.
To cite this abstract in AMA style:
Merriman T, Matsuo H, Takei R, Leask M, Topless R, Shirai Y, Li Z, Cadzow M, Reynolds R, saag k, Fadason T, O'Sullivan J, Dalbeth N, Stamp L, Abhishek A, Doherty M, Roddy E, Jacobsson L, Kapetanovic M, Andrès M, Perez-Ruiz F, Torres Jimenez R, Radstake T, Jansen T, Janssen M, Joosten L, Crisan T, Huizinga T, LIOTE F, Richette P, Bardin T, Pascart T, McCarthy G, Stiburkova B, Tausche A, Uhlig T, Vitart V, Riches P, Ralston S, MacDonald T, Nakayama A, Nakatochi M, Ichida K, Takada T, Lee C, Brown M, Robinson P, Hill C, Choi H, Sumpter N, Merriman M, Phipps-Green A, Wei W, McCormick S, Melander O, Toes R, Ea H, Kurreeman F, Helbert L, Boutin T, Shinomiya N, Bradbury L, Buchanan R, Lester S, Smith M, Rischmueller M, Japan Gout Genomics Consortium (J-Gout) O, Japan Multi-Instl Collab Cohort Study (J-MICC) O, Stahl E, Miner J, Solomon D, Cui J, Giacomini K, Brackman D, Jorgenson E, 23andMe Research Team O, Shringapure S, So A, Okada Y, Li C, Shi Y, Major T. A Genome-Wide Association Analysis of 2,622,830 Individuals Reveals New Pathogenic Pathways in Gout [abstract]. Arthritis Rheumatol. 2022; 74 (suppl 9). https://acrabstracts.org/abstract/a-genome-wide-association-analysis-of-2622830-individuals-reveals-new-pathogenic-pathways-in-gout/. Accessed .« Back to ACR Convergence 2022
ACR Meeting Abstracts - https://acrabstracts.org/abstract/a-genome-wide-association-analysis-of-2622830-individuals-reveals-new-pathogenic-pathways-in-gout/