Session Information
Session Type: Poster Session B
Session Time: 9:00AM-10:30AM
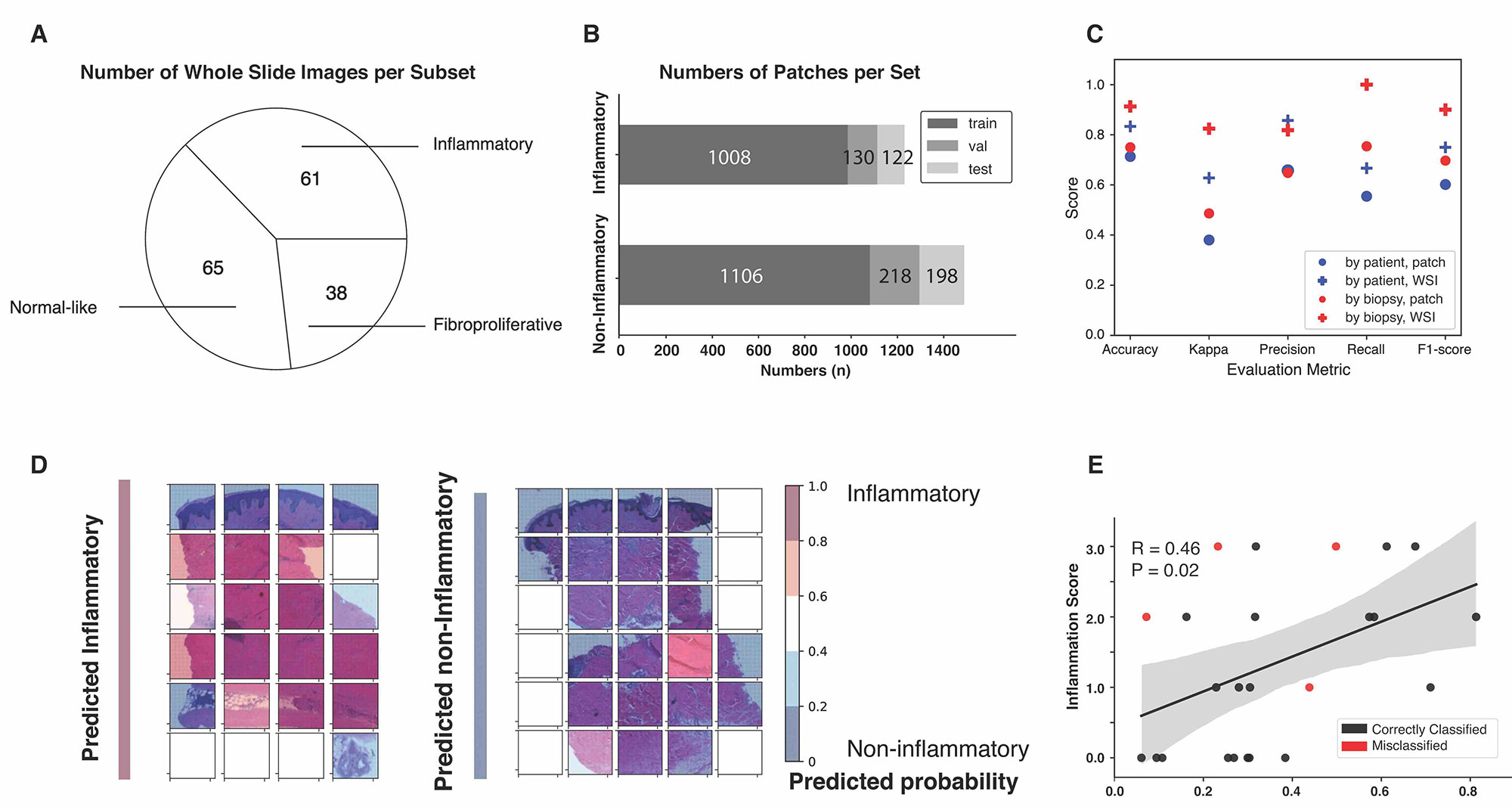
Background/Purpose: Intrinsic gene analyses based on the gene expression profiles in skin have identified four intrinsic molecular subsets among patients with systemic sclerosis (SSc); these have been named the inflammatory, fibroproliferative, limited, and normal-like subsets. SSc participants assigned to the inflammatory subset showed significant improvement in the Abatacept Systemic SclErosis Trial (ASSET). In order to provide a fast and accessible molecular subset predictor, we trained a deep neural network to efficiently identify these participants using the images of hematoxylin and eosin (H&E)-stained skin biopsies.
Methods: Skin tissue, H&E-stained images, and clinical scores were obtained from participants enrolled in the ASSET clinical trial. Image labels were molecular subset calls from a classifier using gene expression from a paired skin biopsy. A sliding window approach was used to generate small patches from whole slide images. Convolutional neural networks (CNNs) were trained on patch images, and patch-level classifications were aggregated to infer the subset of the given biopsy. Visualization for areas of the image that are important for prediction and Spearman correlations between predicted probabilities and clinical scores were conducted for validation. Final prediction for molecular subsets was determined by the combination of individual binary classifiers for the inflammatory, fibroproliferative and normal-like subsets.
Results: CNNs were trained and validated on small patches generated from 140 whole-slide images. The model was evaluated using a hold-out test set of 24 whole-slide images. The final model, which predicts the inflammatory, fibroproliferative and normal-like subsets, identified participants’ molecular subset with 79.17% average accuracy when data sets are split-by-patient. Visualization of patches and predicted decisive regions provided insights on important cell types contributing to the molecular subsets. Predicted molecular subset probabilities were significantly correlated with clinical inflammation and fibrosis scores.
Conclusion: We developed an ensemble neural network classifier that classifies SSc patients into three of the intrinsic molecular subsets found in diffuse cutaneous SSc patients using standard H&E-stained images of skin biopsies. This is less costly than genomic assays to identify these individuals and can be applied to evaluate retrospective data where genomic data were not collected.
To cite this abstract in AMA style:
Yuan Y, Song Q, Harms P, Gudjonsson J, Lafyatis R, Khanna D, Whitfield M. A Deep Neural Network Classifier to Identify Intrinsic Molecular Subsets of Systemic Sclerosis Using Histological Images [abstract]. Arthritis Rheumatol. 2022; 74 (suppl 9). https://acrabstracts.org/abstract/a-deep-neural-network-classifier-to-identify-intrinsic-molecular-subsets-of-systemic-sclerosis-using-histological-images/. Accessed .« Back to ACR Convergence 2022
ACR Meeting Abstracts - https://acrabstracts.org/abstract/a-deep-neural-network-classifier-to-identify-intrinsic-molecular-subsets-of-systemic-sclerosis-using-histological-images/