Session Information
Date: Monday, October 27, 2025
Title: (0978–1006) T Cell Biology & Targets in Autoimmune & Inflammatory Disease Poster
Session Type: Poster Session B
Session Time: 10:30AM-12:30PM
Background/Purpose: MicroRNAs, such as miR-24 and miR-27, co-expressed within the Mirc11 and Mirc22 clusters, orchestrate a regulatory network critical to Th2 cell differentiation and cytokine production, notably Interleukin-4 (IL-4), with emerging roles in immune dysregulation in rheumatic diseases. To probe this network, we developed DeepRNA-Reg, a deep learning-based algorithm leveraging recurrent neural networks for high-fidelity comparative analysis of high-throughput sequencing of RNA isolated by crosslinking immunoprecipitation (HITS-CLIP) datasets.
Methods: The DeepRNA-Reg algorithm utilizes deep learning to predict differentially enriched sites in paired HITS-CLIP data sets, employing a recurrent neural network to infer differential enrichment status across the transcriptome. CD4+ T cells were isolated from spleen and lymph nodes and cultured under Th2 conditions. C57BL/6 male and female age and sex matched mice between 5 to 12 weeks of age were used for all experiments. To generate “miR-23, 24, 27 KO” T cells lacking expression of all Mirc11 cluster miRNAs (miR- 23a, miR-24-2, and miR-27a) and Mirc22 cluster miRNAs (miR-23b, miR-24-1, and miR-27b), we intercrossed previously generated conditionally mutant Mirc22tm1.1Mtm mice and Mirc11tm1.1Mtm mice and with Cd4-Cre transgenic mice (Tg(CD4-cre)1Cwi; 4196, Taconic) to induce Cre-mediated deletion in T cells. Ago2 HITS- CLIP was performed using an anti-Ago2 monoclonal antibody. Lastly, differential protein expression was evaluated in CD4+ T Cells harvested from mice bearing selective microRNA cluster knock-out.
Results: This analysis of miR-24, 27 targets uncovered a suite of transcriptional regulators that are known to affect IL-4 expression, including Gata3 and Ikzf1 but also Rora, Tcf7, Runx1, Bcl11b, Foxo1, Nfat5, Tsc22d3 and Klf11. In addition, this study uncovered the cell surface costimulatory receptor CD28 as a novel target of miR-24 and miR-27. CD28 is a key regulator of T cell activation, differentiation and cytokine production. Flow cytometric analysis of CD28 protein expression confirmed that miR-23,24,27 KO Th2 cells express a larger quantity of CD28 on their cell surface, consistent with direct targeting by miR-24 and/or miR- 27.
Conclusion: These findings demonstrate that DeepRNA-Reg analysis of comparative Ago2 HITS-CLIP data can both confirm prior knowledge and uncover novel nodes in post-transcriptional regulatory networks. In tested data sets, DeepRNA-Reg uncovered novel mediators in the mechanism of microRNA-mediated restraint of type-2 immunity in T-Helper 2 cells, a pattern of immune response implicated in the pathophysiology.
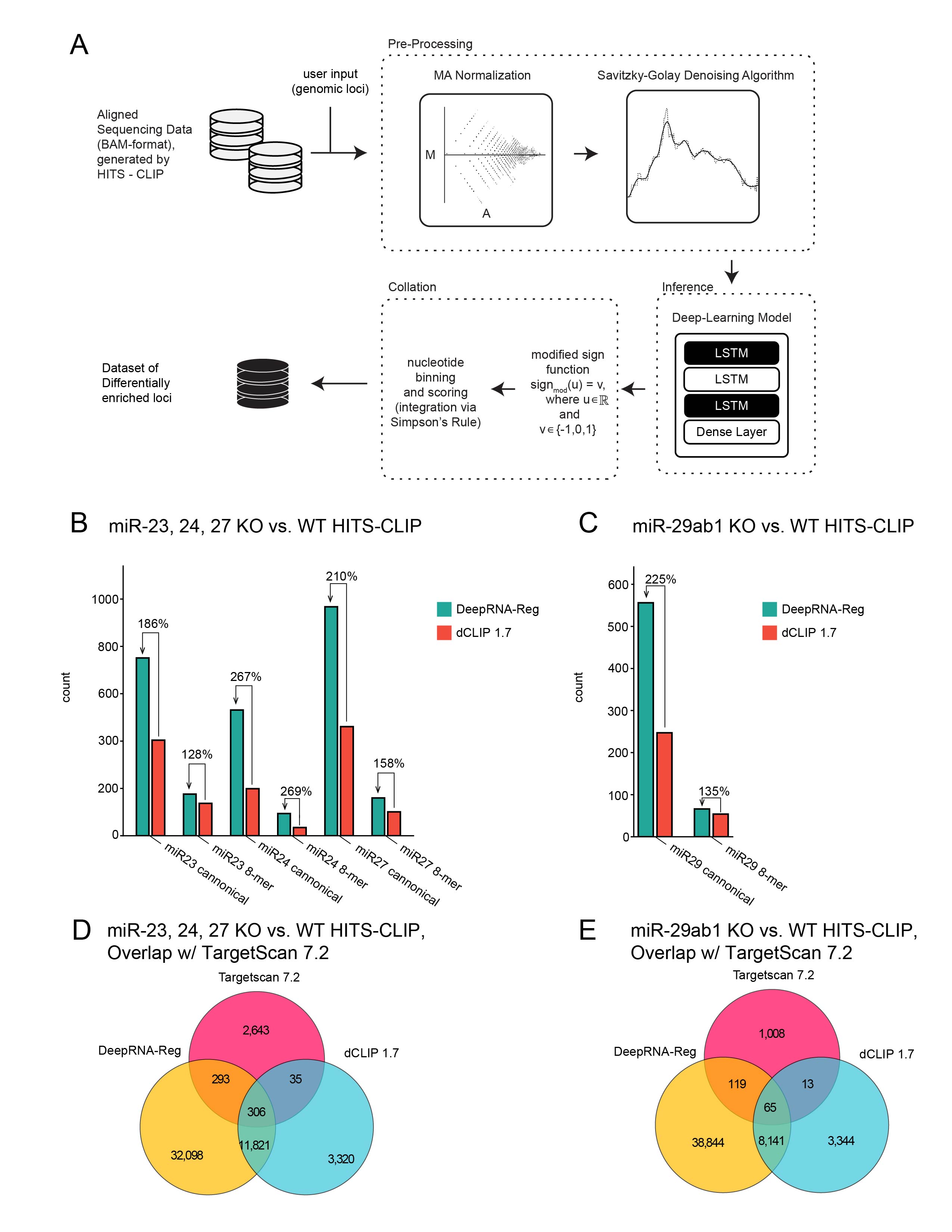 Benchmarking predicted miRNA target site capture.
Benchmarking predicted miRNA target site capture.
(A) Schematic representation of the DeepRNA-Reg algorithm.
(B-C) Seed binding sequence motif abundance in the Th2 differential AHC analysis (B) and the Th17 differential AHC analysis (C) by both DeepRNA-Reg and dCLIP.
(D) Venn diagram quantifying overlap events between Targetscan 7.2 predicted miRNA target sites of miR-23, 24, and 27 and the sites called to be enriched in WT Th2 cells (relative to miR-23,24,27 KO Th2 cells) by both DeepRNA-Reg and dCLIP.
(E) Venn diagram quantifying overlap events between Targetscan 7.2 predicted miRNA target sites of miR-29 and the sites called to be enriched in WT Th17 cells (relative to miR-29 KO Th2 cells) by both DeepRNA-Reg and dCLIP.
.jpg) Interrogating the Il-4 regulatory axis in the T Helper 2 Context.
Interrogating the Il-4 regulatory axis in the T Helper 2 Context.
(A) A gene regulatory network showing the intersection of the IPA knowledge bank of known il-4 regulators and high-confidence DeepRNA-Reg predicted microRNA-24,27 targets. This high-confidence prediction set met the following conditions: a differentially enriched site with a DBE score above the median DBE score for the analysis that contains a canonical motif, with a negative response to the corresponding miRNA mimic between 15% and 85%.
(B) A scatterplot of the degree of gene expression change between the miRNA mimic and control condition for microRNAs 24 and 27 on the horizontal and vertical axes, respectively. This plot represents the set of genes that lie at the intersection of the set of known upstream regulators of interluken-4 within the IPA knowledge-base and the set of high-confidence DeepRNA-Reg predictions of miR-24,27 targets.
(C) A line plot representing the read depth across a section of the Cd28 3’UTR for both the WT and miR-23,24,27 KO condition HITS-CLIP experiments. Highlighted within this plot are two loci enriched in the WT condition relative to the miRNA KO condition, called and scored by DeepRNA-Reg, which overlap canonical seed sequences of miRNAs 24 and 27.
(D) Flow cytometric analysis of CD28 protein expression showing that miR-23,24,27 KO Th2 cells express a larger quantity of CD28 on their cell surface, consistent with direct targeting by miR-24 and/or miR-27
.jpg) Secondary structure of Ago2 binding sites.
Secondary structure of Ago2 binding sites.
(A-C) Heatmaps represent a visualization of the A/U richness (A) and icSHAPE reactivity score – both in vivo (B) and in vitro (C) – for various sites of putative Ago2-miRNA binding targets. The datasets of putative Ago2-miRNA binding sites are the DeepRNA-Reg and dCLIP predicted set of miR-23,24,27 targets provided by the WT Th2 vs. miR-23,24,27 KO Th2 differential AHC analysis, and the set of all Ago2 binding sites called by the algorithm Clipper 2.0 within a dataset of HITS-CLIP performed within WT T-Helper 2 Cells. Below the heatmaps are line plots representing the average signal presented within the heatmaps, collapsed across all predicted binding sites of Ago2.
(D-F) Heatmaps showing the free energy change of dimerization of a rolling 10-bp window surrounding the miRNA seed containing site and the miRNA seed containing region (a 10 bp window centered at the site center) for the set of all Ago2 binding sites called by the algorithm Clipper 2.0 within a dataset of HITS-CLIP performed within WT T-Helper 2 Cells (D), and the DeepRNA-Reg (E) and dCLIP (F) predicted set of miR-23,24,27 targets provided by the WT Th2 vs. miR-23,24,27 KO Th2 differential AHC analysis. Plots located below show the average signal across all sites presented in the heatmap.
To cite this abstract in AMA style:
Sekhon S, Kageyama R, Sprenkle N, Happ H, Wigton E, Pua H, Ansel M. A Deep-Learning Based Approach Uncovers Novel Mediators of Micro-RNA Restraint of Type-2 Immunity [abstract]. Arthritis Rheumatol. 2025; 77 (suppl 9). https://acrabstracts.org/abstract/a-deep-learning-based-approach-uncovers-novel-mediators-of-micro-rna-restraint-of-type-2-immunity/. Accessed .« Back to ACR Convergence 2025
ACR Meeting Abstracts - https://acrabstracts.org/abstract/a-deep-learning-based-approach-uncovers-novel-mediators-of-micro-rna-restraint-of-type-2-immunity/
