Session Information
Date: Wednesday, October 29, 2025
Title: Abstracts: Systemic Lupus Erythematosus – Diagnosis, Manifestations, & Outcomes III (2645–2650)
Session Type: Abstract Session
Session Time: 9:45AM-10:00AM
Background/Purpose: Systemic lupus erythematosus (SLE) is a heterogenous autoimmune disease with significant variability in clinical presentation. DNA methylation has been associated with lupus risk and heterogeneity. In order to identify biologically relevant molecular subtypes, we leveraged genome-wide methylation profiles to cluster patients and evaluate associations between methylation-based clusters and SLE clinical characteristics.
Methods: We analyzed methylation profiles from 1,882 multi-ancestry SLE patients. Clinical data was collected at enrollment and time of peripheral blood collection. Genome-wide methylation profiles were generated using Illumina’s EPICv2 BeadChip (n= 937,690 CpG sites). Normalization and quality control were performed using minfi. CpGs with high detection p-values and cross-reactive probes were excluded, and missing methylation values imputed using methyLImp2. We selected the top 5% most variable CpGs (~60,000) by beta value standard deviation and performed unsupervised k-means clustering using ConsensusClusterPlus across multiple CpG sets (n = 5,000; 10,000; 25,000; 60,000 CpGs). The most distinct clustering was observed with 60,000 CpGs and k=4. Differentially methylated positions (DMPs) by cluster were identified using limma, with beta values used for coefficient interpretation and M values to determine statistical significance. Models adjusted for genetic principal components, Houseman estimated blood cell proportions, batch, age, disease duration, sex. DMPs with FDR q< 0.05 were considered statistically significant.
Results: We identified 4 distinct clusters based on methylation beta values (Figure 1). Clusters 1 and 2 reflected milder disease with discoid rash (p=0.002); Cluster 3 exhibited older age at enrollment (p=0.001); Cluster 4 was associated with more severe disease as reflected by younger age of diagnosis (p=0.006), serositis (p< 0.001), nephritis (p< 0.001), higher proportion of males (p=0.006) and hematologic abnormalities (p< 0.001). 2,569 CpGs with FDR q< 0.05 were identified across all cross-comparisons between clusters (Figure 2). Significant CpGs annotated to ZFP36L1 (an RNA-binding protein that plays a role in post-transcriptional gene regulation), DOCK9 (involved in intracellular signaling networks), and NEK8 (protein kinase involved in cell cycle regulation, DNA damage response, and mitosis). Of the significant CpG sites, the absolute change in methylation between clusters ranged from -4.6% to 3.8 %. A majority of these were hypomethylated; 267 were located within genes, and 322 were in promoter regions. Gene set enrichment analysis (GSEA) revealed cluster-specific enrichment: Cluster 1 in leukocyte cell adhesion (FDR q=0.025) and leucocyte activation involved in immune response (FDR q=0.025); Cluster 2 in positive regulation of MAPK (FDR q=0.0003); Cluster 3 in neutrophil degranulation (FDR q=0.001); and Cluster 4 in pattern specification (FDR q=1e-5) and cell adhesion (FDR q=1e-5).
Conclusion: We identified 2,569 differentially methylated CpGs across 4 distinct SLE clusters. These results support the potential of DNA methylation profiling to reveal molecular subtypes of SLE and improve risk stratification.
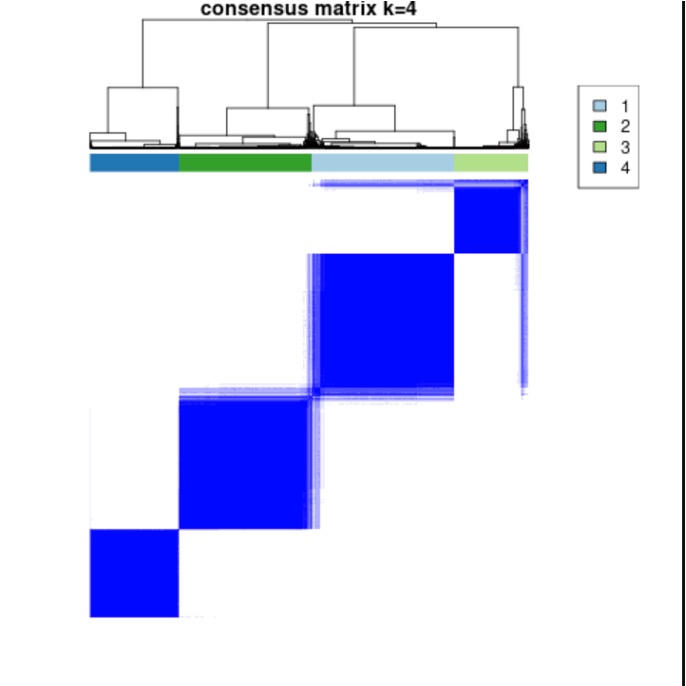 Figure 1. Consensus Plot (k=4) of Patient Clusters based off methylation beta values.
Figure 1. Consensus Plot (k=4) of Patient Clusters based off methylation beta values.
.jpg) Figure 2. Volcano Plot of Significant CpGs shared across all cross-clusters.
Figure 2. Volcano Plot of Significant CpGs shared across all cross-clusters.
To cite this abstract in AMA style:
Nelson M, Horton M, Nititham J, Yazdany J, Dall'Era M, Lanata C, Criswell L. DNA methylation-based clustering reveals clinically distinct subtypes of systemic lupus erythematosus [abstract]. Arthritis Rheumatol. 2025; 77 (suppl 9). https://acrabstracts.org/abstract/dna-methylation-based-clustering-reveals-clinically-distinct-subtypes-of-systemic-lupus-erythematosus/. Accessed .« Back to ACR Convergence 2025
ACR Meeting Abstracts - https://acrabstracts.org/abstract/dna-methylation-based-clustering-reveals-clinically-distinct-subtypes-of-systemic-lupus-erythematosus/
