Session Information
Session Type: Poster Session C
Session Time: 10:30AM-12:30PM
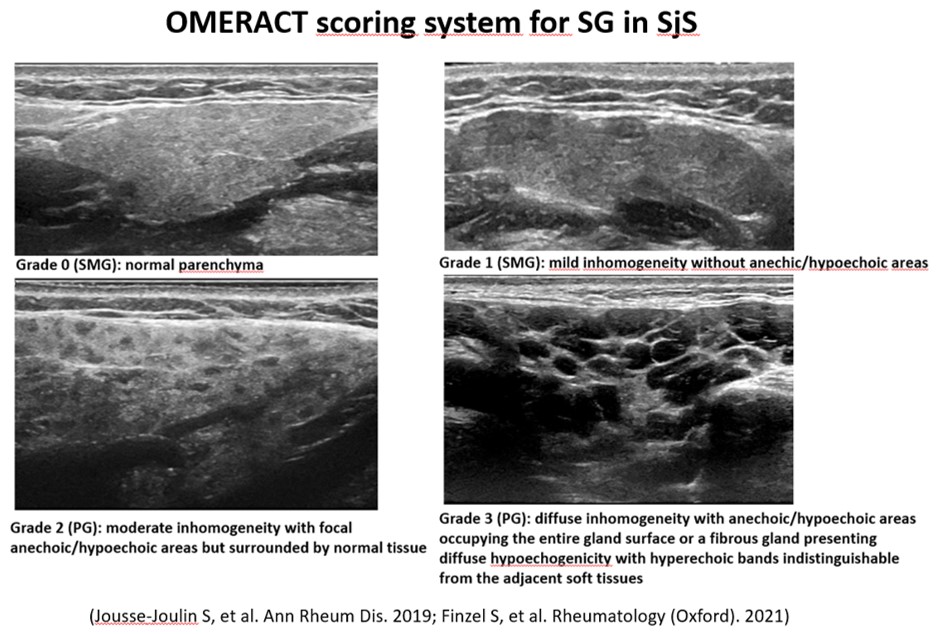
Background/Purpose: To develop a deep learning model for classifying ultrasound images of salivary glands (SG) [parotid (PG) and submandibular (SMG)] based on the Outcome Measures in Rheumatology (OMERACT) 0-3 semiquantitative scoring system for B-mode ultrasound-assessed parenchymal abnormalities in SG in Sjögren syndrome (SjS) (Figure 1)
Methods: For training, we used 3 datasets of SG (PG and SMG) ultrasound images representative of the OMERACT scores: 1) 225 images from 150 suspected or confirmed SjS patients acquired with different ultrasound systems across 4 European centers (https://www.frontiersin.org/articles/10.3389/fmed.2020.581248/full); 2) 80 images from 20 SjS patients provided by a European center; 3) Of 39 SjS patients and 10 controls, 6 images of each acquired by 2 experts with 2 systems (a high-end and a wireless handheld device) in a European center (1176 images). We used a ResNet18 architecture, which was pre-trained on ImageNet and then fine-tuned for this task. The model was trained hierarchically: initially classifying broad categories (scores 0/1 vs. 2/3) and subsequently refining the classification for the specific stages of the disease. 70% of the images were used for training, 15% for validation, and 15% for test. For training, we used a dataset of ultrasound images along with labels indicating the grade of SG involvement. We employed standard deep learning techniques such as data augmentation (e.g., random flipping and brightness adjustments) to increase the model’s robustness and generalization capabilities and a step learning rate scheduler to optimize the training process. Precision (correct classifications per predicted class), recall (correct classifications per actual class), and F1-score (harmonic mean of precision and recall) were calculated for each grading class.
Results: The overall performance of the model for grading classes is shown in Table 1. The broad model achieved an accuracy of 90% in distinguishing between 0/1 and 2/3 grades. However, the more specific models, fine-tuned for the 0/1 and 2/3 categories, had lower accuracy (approximately 65% for each). This reflects the increased difficulty of fine-grained classification. The confusion matrix (Figure 2) showed that the model performed well in classifying SG with grade 3 but faced challenges with intermediate categories (class 1 and class 2). The network showed difficulty discriminating between grade 0 and 1; on retrospective review of the ultrasound labels, we noted that this boundary is visually subtle and prone to greater inter-observer variability, which likely contributes to the model’s confusion between these two lowest scores.
Conclusion: Our results showed the potential of deep learning models, specifically convolutional neural networks, to classify SG ultrasound images based on parenchymal involvement, offering a promising tool for SjS diagnosis. While the broad classification model performed very well, the fine-tuned models, which differentiate between more specific disease stages, still faced challenges. Future work should refine the fine-tuned models with additional data, allowing more complex architecture or domain-specific adjustments.
To cite this abstract in AMA style:
Olivas O, García-Sevilla M, Cubero L, Pascau J, Naredo E. Performance of Artificial Intelligence-Based Salivary Gland Ultrasound in Sjögren Syndrome [abstract]. Arthritis Rheumatol. 2025; 77 (suppl 9). https://acrabstracts.org/abstract/performance-of-artificial-intelligence-based-salivary-gland-ultrasound-in-sjogren-syndrome/. Accessed .« Back to ACR Convergence 2025
ACR Meeting Abstracts - https://acrabstracts.org/abstract/performance-of-artificial-intelligence-based-salivary-gland-ultrasound-in-sjogren-syndrome/

.jpg)
.jpg)