Session Information
Session Type: Poster Session C
Session Time: 10:30AM-12:30PM
Background/Purpose: Accurate identification of patients with psoriasis from large electronic health record (EHR) databases is crucial for conducting robust real-world research. While EHRs offer an efficient resource, traditional identification methods are often laborious. Machine learning (ML) can present a promising high throughput approach with accuracy. The objective of this study was to develop and validate machine learning algorithms using EHR data to identify patients with psoriasis.
Methods: We used data from Vanderbilt University Medical Center’s de-identified EHR database, the Synthetic Derivative (SD), which contains longitudinal clinical data from approximately 3.5 million subjects. Adult patients (≥18 years) were initially screened based on potential psoriasis indicators like International Classification of Diseases (ICD) codes or keywords. Non-overlapping random samples formed the training (Nf200) and validation (Nf300) sets. A true psoriasis case was defined through manual chart review as documented confirmatory or probable diagnosis by a dermatologist or rheumatologist, or a positive skin biopsy. Potential machine learning features included ICD-9/10 codes for psoriasis and related conditions, provider specialty associated with codes (dermatology, rheumatology), problem list keywords, and use of relevant medications (e.g., topical treatments, systemic therapies, biologics). Three ML models (logistic regression, random forest, XGBoost) were developed on the training set, with the random forest model undergoing hyperparameter tuning via repeated 10-fold cross-validation. Models were tested on the independent validation set, assessing performance using F1-score, sensitivity, positive predictive value (PPV), area under the receiver operating characteristic curve (AUC), and accuracy.
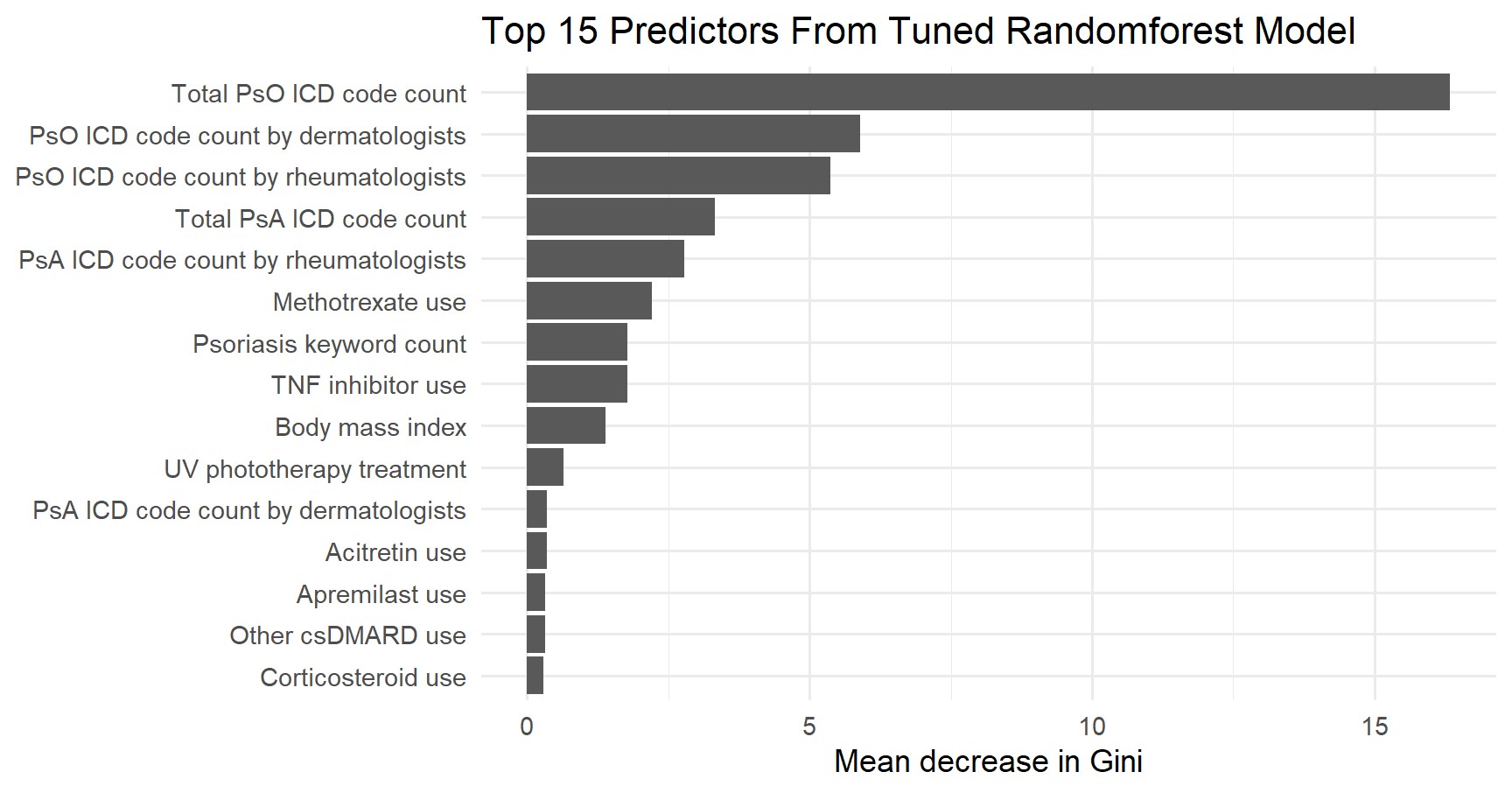
Results: All three ML models demonstrated strong performance identifying psoriasis patients in the validation set. Tuned random forest model achieved the highest F1-score (0.757), with an AUC of 0.871, sensitivity of 0.791, and PPV of 0.725 (Table 1, Figure 1). Feature importance analysis confirmed that ICD codes related to psoriasis diagnoses documented by dermatologists and rheumatologists, overall psoriasis visit codes, and specific medication usage patterns were among the most significant predictors of a true psoriasis diagnosis (Figure 2).
Conclusion: Machine learning algorithms, particularly tuned random forest models, leveraging readily available EHR data could provide an accurate and efficient method for identifying psoriasis patients comparable to true diagnosis made by dermatologist or rheumatologist or with skin biopsy. Such validated algorithms represent valuable tools for researchers seeking to assemble large, real-world cohorts of patients with psoriasis enabling studies on disease epidemiology, treatment effectiveness, and patient outcomes.
 Comparison of Performances Among Three Different Machine Learning Models
Comparison of Performances Among Three Different Machine Learning Models
.jpg) Receiver Operating Curves with Area Under Curve for Random Forest Model in Training and Validation Sets
Receiver Operating Curves with Area Under Curve for Random Forest Model in Training and Validation Sets
.jpg) Top 15 Predictors of a True Psoriasis Diagnosis
Top 15 Predictors of a True Psoriasis Diagnosis
To cite this abstract in AMA style:
Poudel D, Crofford L, Fortier A, Wheless L, Sellyn G, Sherpa G, Ogdie A, Karmacharya P. Developing Machine Learning Algorithm in Electronic Health Record to Accurately Identify Psoriasis Patients [abstract]. Arthritis Rheumatol. 2025; 77 (suppl 9). https://acrabstracts.org/abstract/developing-machine-learning-algorithm-in-electronic-health-record-to-accurately-identify-psoriasis-patients/. Accessed .« Back to ACR Convergence 2025
ACR Meeting Abstracts - https://acrabstracts.org/abstract/developing-machine-learning-algorithm-in-electronic-health-record-to-accurately-identify-psoriasis-patients/
