Session Information
Date: Tuesday, October 28, 2025
Title: (1830–1854) Systemic Lupus Erythematosus – Etiology and Pathogenesis Poster
Session Type: Poster Session C
Session Time: 10:30AM-12:30PM
Background/Purpose: Systemic lupus erythematosus (SLE) significantly increases the risk of premature atherosclerosis, contributing to long-term morbidity and mortality. Traditional cardiovascular risk factors do not fully explain this phenomenon. Immunologically driven endothelial injury is proposed as a key mechanism. This study integrates transcriptomic and immune cell profiling data to identify molecular and cellular signatures associated with subclinical atherosclerosis in SLE.
Methods: Transcriptomic datasets GSE154851 and GSE110174 were obtained from the Gene Expression Omnibus. GSE154851 includes patients with increased carotid intima-media thickness (CIMT), while GSE110174 includes SLE patients without CIMT and healthy controls. Gene set enrichment analysis (GSEA) was conducted to identify genes linked to coronary atherosclerosis. Protein-protein interaction (PPI) networks were analyzed using Cytoscape, with hub genes (HGs) identified through five centrality algorithms. Immune cell fractions (22 types) were estimated via CIBERSORTx. A two-way ANOVA with FDR correction was used to compare cell populations across groups, and logistic regression assessed associations with CIMT status.
Results: Nineteen genes were significantly enriched in the atherosclerosis phenotype, including ACTA2, ABCA1, MYH11, APOB, PCSK9, PPARG, TGFBR1, and SMAD3. Immune cell profiling revealed significant differences in CD8+ T cells, CD4+ naïve and memory subsets, NK cells, monocytes, M0 macrophages, and neutrophils. CD8+ T cells, CD4+ naïve T cells, and resting NK cells were negatively associated with CIMT, whereas CD4+ memory resting T cells, monocytes, and M0 macrophages showed positive associations (all p < 0.001). Diagnostic accuracy was supported by ROC curve analysis, highlighting monocytes and CD4+ memory T cells as potential biomarkers.
Conclusion: This study uncovers specific immune cell subsets and gene networks linked to atherosclerosis in SLE. The findings emphasize the contribution of innate and adaptive immune dysregulation in vascular pathology and support further exploration of immune-based biomarkers to stratify cardiovascular risk in lupus.
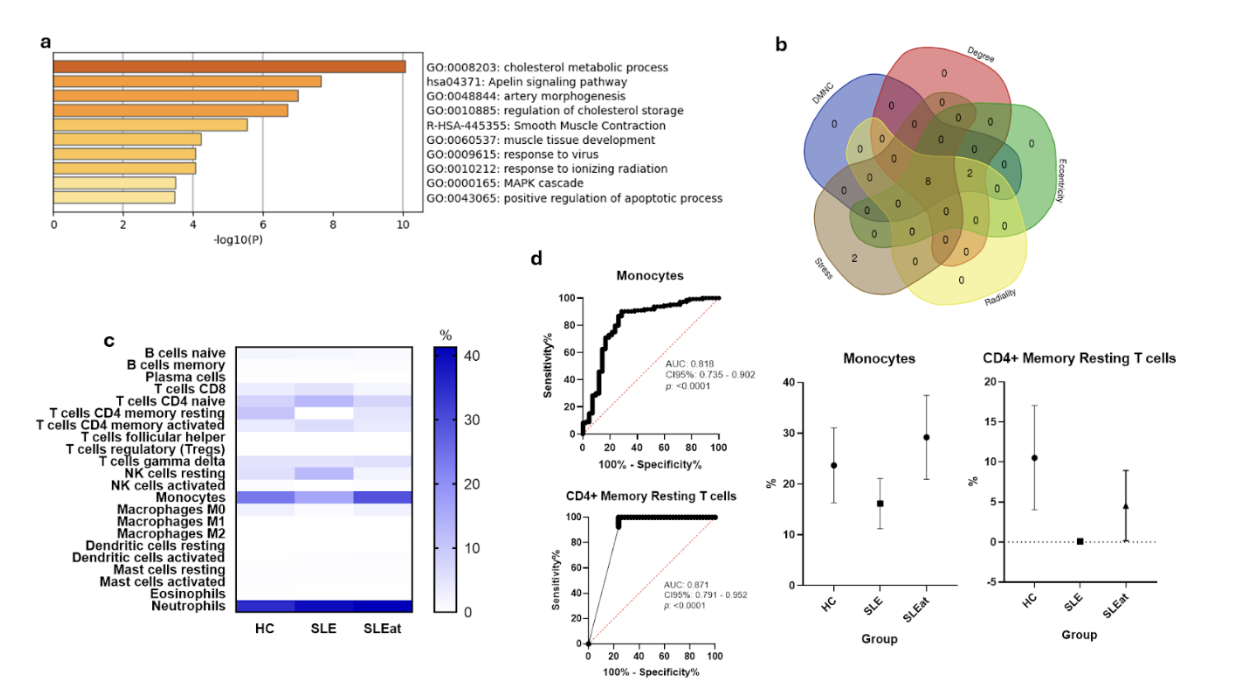 Figure 1. Immune profiling and diagnostic relevance of hub genes in SLE-related atherosclerosis. (a) Gene set enrichment analysis of transcripts linked to coronary atherosclerosis in individuals with systemic lupus erythematosus (SLE). (b) Common hub genes determined by the intersection of five centrality-based algorithms. (c) Relative abundance of 22 immune cell subsets in healthy controls (HC), SLE patients without carotid intima-media thickening (CIMT), and SLE patients with increased CIMT (SLEat). (d) Receiver operating characteristic (ROC) curves showing the predictive performance of monocyte counts and resting CD4+ memory T cells for identifying subclinical atherosclerosis in the SLE cohort.
Figure 1. Immune profiling and diagnostic relevance of hub genes in SLE-related atherosclerosis. (a) Gene set enrichment analysis of transcripts linked to coronary atherosclerosis in individuals with systemic lupus erythematosus (SLE). (b) Common hub genes determined by the intersection of five centrality-based algorithms. (c) Relative abundance of 22 immune cell subsets in healthy controls (HC), SLE patients without carotid intima-media thickening (CIMT), and SLE patients with increased CIMT (SLEat). (d) Receiver operating characteristic (ROC) curves showing the predictive performance of monocyte counts and resting CD4+ memory T cells for identifying subclinical atherosclerosis in the SLE cohort.
To cite this abstract in AMA style:
Martinez-Canales R, Ortiz-Rios A, Avalos-Garcia B, Galindo-Calvillo E, Salinas-Carmona M, Macias-Segura N, Perez-Barbosa L, Galarza-Delgado D, Skinner-Taylor C. Immune Cell Profiles and Transcriptomic Signatures of Atherosclerosis in Systemic Lupus Erythemathosus [abstract]. Arthritis Rheumatol. 2025; 77 (suppl 9). https://acrabstracts.org/abstract/immune-cell-profiles-and-transcriptomic-signatures-of-atherosclerosis-in-systemic-lupus-erythemathosus/. Accessed .« Back to ACR Convergence 2025
ACR Meeting Abstracts - https://acrabstracts.org/abstract/immune-cell-profiles-and-transcriptomic-signatures-of-atherosclerosis-in-systemic-lupus-erythemathosus/

.jpg)