Session Information
Date: Tuesday, October 28, 2025
Title: (1830–1854) Systemic Lupus Erythematosus – Etiology and Pathogenesis Poster
Session Type: Poster Session C
Session Time: 10:30AM-12:30PM
Background/Purpose: In collaboration with the AMP-RA/SLE network, we identified disease-associated macrophages (D-Macs) in kidney biopsies from 155 patients with active lupus nephritis (LN) and 30 controls, using single-cell RNA sequencing of ~25,000 myeloid cells (Fig. 1A, B). D-Macs comprised several macrophage subsets and expressed an injury-associated gene program, including TREM2, CD63, CTSD, APOE, SPP1, GPNMB, CD9, and FABP5 (Fig. 1C), which are conserved across monocytes and macrophages in other injured tissues. D-Macs showed positive correlation with the ISN/RPS activity index and were more prevalent in proliferative and mixed histological classes (III/IV±V) compared to the pure membranous class (V) or healthy individuals (not shown). In this follow-up study, we explored the origins and significance of D-Macs in proliferative/mixed LN.
Methods: We analyzed kidney biopsies from LN patients and controls using single-cell RNA sequencing to identify distinct cell populations. Spatial transcriptomics was performed with a custom gene panel to map myeloid subtypes in FFPE kidney samples. We also examined transcriptomic responses to putative nephritic glomerular factors plus M-CSF in healthy CD14 monocytes using flow cytometry and RNA sequencing.
Results: Bioinformatic analysis suggested that D-Macs likely differentiate in situ from infiltrating monocytes or resident macrophages, indicating a process of “convergent differentiation” where blood and tissue myeloid cells adopt similar transcriptional profiles in response to factors from diseased kidneys (Fig. 1D-G). To gain further insight into D-Mac differentiation and function, we mapped each D-Mac subset alongside other myeloid populations across renal compartments and in relation to lesions in FFPE sections from 6 LN patients with proliferative or mixed histologic classes (mean ISN/RPS activity = 13.5; S.D = 5.8) and 2 controls. Certain D-Mac subsets were statistically enriched in glomeruli with proliferative lesions (Glomerular D-Macs), while others were mainly found in the tubulointerstitium (Tubulointerstitial D-Macs) (Fig. 2), suggesting that distinct D-Mac subsets may differentiate from different precursors in a compartment-specific manner.To determine whether factors from nephritic glomeruli drive D-Mac differentiation from infiltrating CD14+ or CD16+ monocytes, we screened 40 inflammatory mediators identified through single-cell data, spatial transcriptomics, histology, and literature review. Results showed that several glomerular factors—such as immune complexes with resiquimod, cell debris, and TGF-β—promoted the induction of D-Mac-like states, including an injury-associated gene program similar to that observed in patients, from both CD14+ (Fig. 3) and CD16+ (not shown) monocyte precursors.
Conclusion: Our findings highlight key glomerular mediators in proliferative lupus nephritis that may promote D-Mac differentiation from infiltrating monocytes. They support the idea that distinct monocyte states respond similarly to nephritic factors, converging on a shared cellular phenotype. Ongoing studies are focused on elucidating the functional roles of these cells.
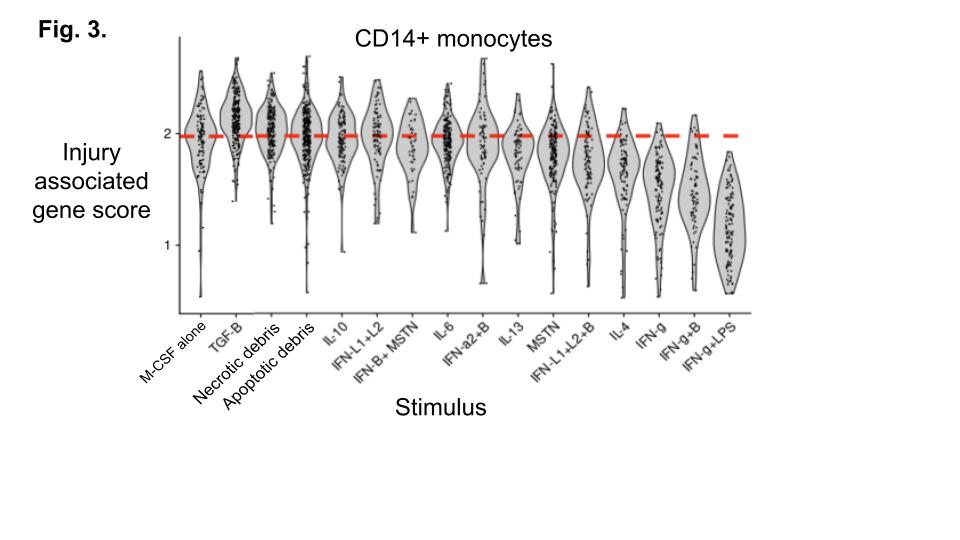 Fig. 1. (A-C) UMAP of single-cell analysis of myeloid cells labeled by clusters (A), case vs. controls (B), or average expression of injury associated genes. Black dotted line outlines the D-Mac clusters. (D-G) Assessing the feasibility of differentiation of monocytes and RMs into DMac clusters. The shortest paths from each putative origin into each DMac cluster is shown based on the results of trajectory analysis using partition based graph abstraction (PAGA) (PMID 30890159). Edges colored in red area also supported by the colocalization analysis presented below in Fig. 3. Paths originating from (D) CD16+ nonclassical monocytes (cluster M2); (E) CD14+ classical monocytes (cluster M3); (F) Lyve1+ RMs (cluster M9); (G) Lyve1- RMs (cluster M0).
Fig. 1. (A-C) UMAP of single-cell analysis of myeloid cells labeled by clusters (A), case vs. controls (B), or average expression of injury associated genes. Black dotted line outlines the D-Mac clusters. (D-G) Assessing the feasibility of differentiation of monocytes and RMs into DMac clusters. The shortest paths from each putative origin into each DMac cluster is shown based on the results of trajectory analysis using partition based graph abstraction (PAGA) (PMID 30890159). Edges colored in red area also supported by the colocalization analysis presented below in Fig. 3. Paths originating from (D) CD16+ nonclassical monocytes (cluster M2); (E) CD14+ classical monocytes (cluster M3); (F) Lyve1+ RMs (cluster M9); (G) Lyve1- RMs (cluster M0).
.jpg) Fig. 2. Specific D-Mac subsets localize to glomeruli or tubulointerstitial regions of tissue. (A) Top panel shows example biopsy from patient with cass IV+V, activity 16/24 and chronicity 5/12. Bottom panel shows (B) areas of kidney in our analysis and (C) quantification of enrichment (red), depletion (blue), and statistical significance (asteriks) across indicated areas from 6 patients and 2 controls.
Fig. 2. Specific D-Mac subsets localize to glomeruli or tubulointerstitial regions of tissue. (A) Top panel shows example biopsy from patient with cass IV+V, activity 16/24 and chronicity 5/12. Bottom panel shows (B) areas of kidney in our analysis and (C) quantification of enrichment (red), depletion (blue), and statistical significance (asteriks) across indicated areas from 6 patients and 2 controls.
.jpg) Fig. 3. CD14 monocytes were stimulated with M-CSF alone or with the indicated factors and subject to single cell RNA-seq to measure the average expression of the injury associated score (genes in Fig. 1C).
Fig. 3. CD14 monocytes were stimulated with M-CSF alone or with the indicated factors and subject to single cell RNA-seq to measure the average expression of the injury associated score (genes in Fig. 1C).
To cite this abstract in AMA style:
Hoover P, Leavitt R, Buyon J, Anolik J, Barnas J, James J, Guthridge J, Petri M, Diamond B, Raychaudhuri S, Hacohen N, Davidson A, Arazi A. Disease-Associated Macrophages Express an Injury-Associated Gene Program and Localize to Distinct Compartments in Proliferative and Mixed Histologic Classes of Lupus Nephritis [abstract]. Arthritis Rheumatol. 2025; 77 (suppl 9). https://acrabstracts.org/abstract/disease-associated-macrophages-express-an-injury-associated-gene-program-and-localize-to-distinct-compartments-in-proliferative-and-mixed-histologic-classes-of-lupus-nephritis/. Accessed .« Back to ACR Convergence 2025
ACR Meeting Abstracts - https://acrabstracts.org/abstract/disease-associated-macrophages-express-an-injury-associated-gene-program-and-localize-to-distinct-compartments-in-proliferative-and-mixed-histologic-classes-of-lupus-nephritis/
