Session Information
Session Type: Poster Session C
Session Time: 10:30AM-12:30PM
Background/Purpose: Systemic juvenile idiopathic arthritis (sJIA) is an autoinflammatory disorder characterized by systemic immune dysregulation. However, reliable biomarkers to predict its unpredictable disease course are still lacking. Identifying immune cell subsets and molecular drivers of disease progression is crucial for improving prognosis and developing targeted therapies.
Methods: We performed comprehensive immunophenotypic profiling of PBMCs from sJIA patients across five clinical centers. Deconvolution analysis combined with clinical follow-up in the Chongqing cohort was used to assess the prognostic value of key subpopulations. In vivo validation was performed using UBE2D1 knockout mice. A cross-disease single-cell reference was developed to compare sJIA-associated immune subsets with those found in other inflammatory diseases.
Results: We identified a novel CD14+CXCL10+monocyte subset distinguished by a unique transcriptomic signature enriched in immune regulatory genes. Notably, flow cytometry revealed that this subset was significantly elevated during the active phase of sJIA and decreased during remission, demonstrating a strong correlation with disease activity. In vitro inflammatory stimulation promoted the differentiation of monocytes into the CXCL10+phenotype. But, this process was markedly impaired in UBE2D1-deficient mice, which exhibited lower expression of CXCL10 mediators, underscoring the pivotal role of UBE2D1 as a central driver of this inflammatory program. Finally, by comparing this subset with other JIA subtypes and inflammation-related diseases, we found that it exhibits a distinct expression pattern in sJIA.
Conclusion: Our findings suggest that UBE2D1-driven CD14+CXCL10+monocyte play a key role in sJIA pathogenesis and may serve as both a biomarker and therapeutic target for disease monitoring and intervention.
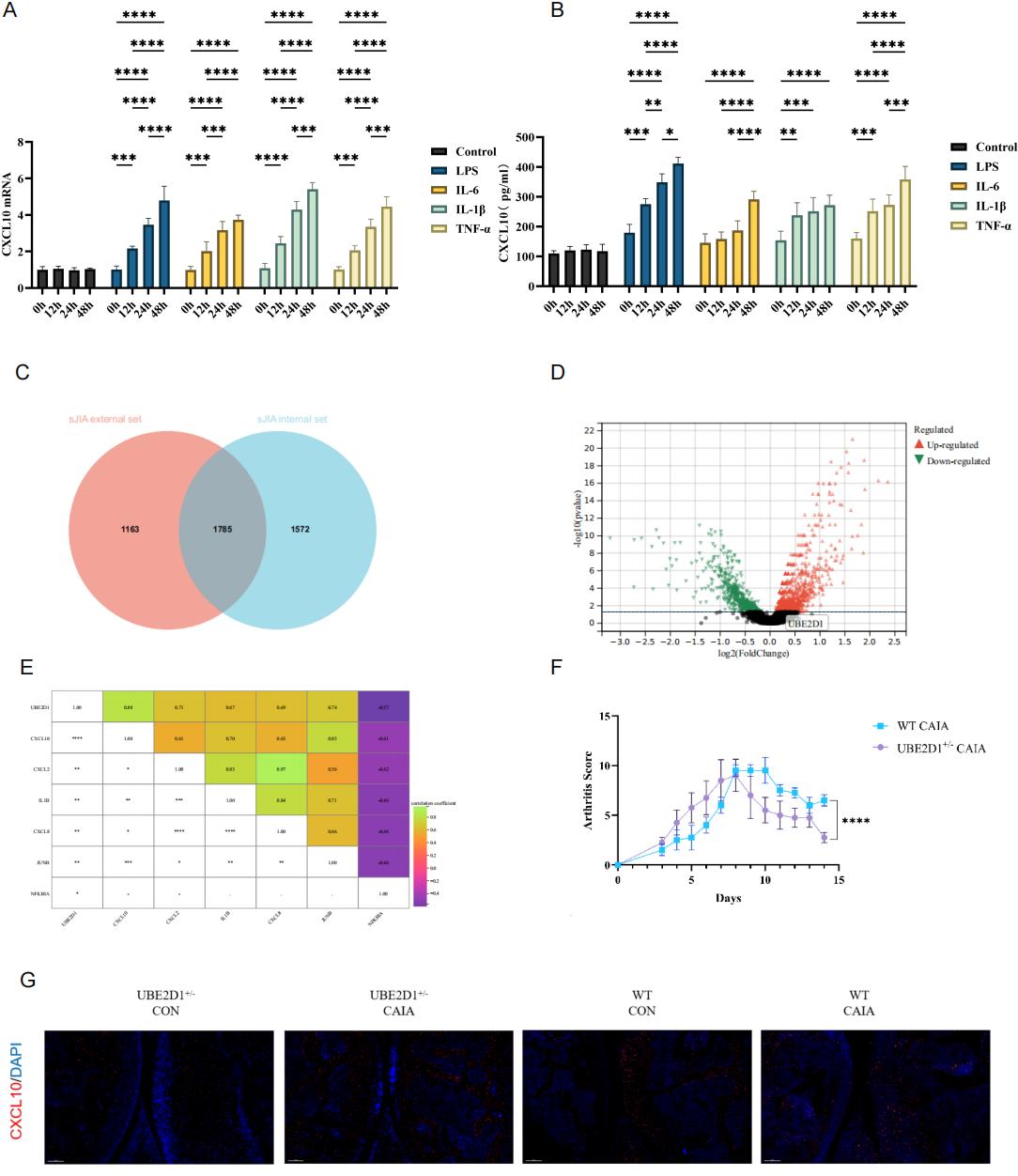 Figure 1: Exploration of CD14+CXCL10+Monocyte Function
Figure 1: Exploration of CD14+CXCL10+Monocyte Function
(A)Exploration of Biological Functions of CD14+CXCL10+Monocyte. (B) Violin plot showing CD14+CXCL10+monocyte levels across pJIA, ERA, oJIA, controls, and sJIA. (C) ROC curve illustrating the diagnostic performance of CD14+CXCL10+monocyte for distinguishing sJIA. (D) Differential expression of monocyte subpopulations in sJIA patients with and without remission. (E) Dot plot showing the Spearman’s correlations of CD14+CXCL10+monocyte and clinical features across the Chongqing Clinical Dataset (*p < 0.05, **p < 0.01, ***p < 0.001, and ****p < 0.0001). (F) Flow cytometry analysis of CD14+CXCL10+monocyte in peripheral blood of sJIA patients and controls. Left: Representative flow cytometry plots showing the percentage of CD14+CXCL10+monocyte in PBMCs from sJIA patients (top row) and controls (bottom row). Top right: Quantification shows that the proportion between sJIA and controls. Bottom right: Longitudinal analysis in the progression of sJIA disease.
.jpg) Figure 2: Exploration of CD14+CXCL10+Monocyte Core Network
Figure 2: Exploration of CD14+CXCL10+Monocyte Core Network
(A-B) Expression of CXCL10 gene and protein in THP-1 cells under different inflammatory stimuli in vitro. (C) Overlapping DEG genes between sJIA internal and external datasets. (D) DEG analysis in post-treatment sJIA and control. (E) Dot plot showing the Spearman’s correlations of UBE2D1 and feature genes (*p < 0.05, **p < 0.01, ***p < 0.001, and ****p < 0.0001). (F) Time-course plot showing arthritis scores over 15 days: UBE2D1⁺/⁻ CAIA mice and WT CAIA mice (****p < 0.0001). (G) Representative multiplex immunofluorescence staining images of joint tissues from four experimental groups: UBE2D1⁺/⁻ CON, UBE2D1⁺/⁻ CAIA, WT CON, and WT CAIA.
.jpg) Figure 3: CD14+CXCL10+ monocyte Show Disease-Specific in Kawasaki Disease, BLAU, sepsis, and sJIA
Figure 3: CD14+CXCL10+ monocyte Show Disease-Specific in Kawasaki Disease, BLAU, sepsis, and sJIA
(A) CD14+CXCL10+ monocyte enriched in sepsis samples. (B) CD14+CXCL10+ monocyte enriched in Kawasaki Disease (KD samples. (C) CD14+CXCL10+ monocyte enriched in BLAU samples. (D) CD14+CXCL10+ monocyte enriched in BLAU samples. (E) Ratio of observed to expected proportions of CD14+CXCL10+ monocyte across four disease. (F) Violin plots displaying UBE2D1 expression levels in CD14+CXCL10+ monocyte from each disease cohort.
To cite this abstract in AMA style:
Tang X, Luo Q, Yang J, Yu H, Wu X, Luo X, Yang X, Zhang Z, An Y, Zhao X, Song H. CD14+CXCL10+Monocytes Remodel the Peripheral Immune Network in sJIA via UBE2D1-Driven Inflammatory Programming [abstract]. Arthritis Rheumatol. 2025; 77 (suppl 9). https://acrabstracts.org/abstract/cd14cxcl10monocytes-remodel-the-peripheral-immune-network-in-sjia-via-ube2d1-driven-inflammatory-programming/. Accessed .« Back to ACR Convergence 2025
ACR Meeting Abstracts - https://acrabstracts.org/abstract/cd14cxcl10monocytes-remodel-the-peripheral-immune-network-in-sjia-via-ube2d1-driven-inflammatory-programming/
