Session Information
Session Type: Poster Session C
Session Time: 10:30AM-12:30PM
Background/Purpose: Childhood-onset lupus is generally associated with a more severe disease course than adult-onset lupus. DNA methylation alterations are known to play a key role in lupus pathogenesis. While we previously showed that childhood-onset lupus is linked to a higher genetic risk burden compared to adult-onset disease, the epigenetic landscape of childhood-onset lupus remains largely unexplored. This study aimed to characterize DNA methylation changes in childhood-onset lupus.
Methods: A total of 64 patients with childhood-onset lupus and 47 healthy controls were included in this study, along with an independent validation cohort of 38 additional patients. DNA was isolated from PBMCs in the study cohort and from whole blood in the validation cohort to assess DNA methylation using the Infinium MethylationEPIC v2.0 array (Illumina). Quality controls and statistical analyses were performed using minfi and limma R packages. Methylation differences among groups were tested through linear regression, adjusting for age, sex, medication use, and cell subset compositions. Differences in clinical manifestations were assessed using Fisher’s exact tests. Gene ontology enrichment analyses were performed with Metascape and GREAT.
Results: Case-control differential DNA methylation analysis revealed significant hypomethylation in interferon-regulated genes in lupus, such as DTX3L, PARP9, IFI44L, and MX1. The enrichment analysis confirmed the presence of type I interferon signature-related biological processes, consistent with adult-onset lupus. Hypomethylation in genes related to B cell activation and cellular senescence was correlated with more active disease, as measured by SLEDAI scores. K-means clustering analysis of patients based on DNA methylation patterns identified 3 distinct lupus clusters. Cluster 1 was characterized by the enrichment of hypomethylated genes involved in cell adhesion and response to growth factor pathways; Cluster 2 exhibited hypomethylation in genes related to regulation of cell differentiation and cell fate determination; and Cluster 3 was enriched in response to oxidative stress and Rap1 signaling pathway in hypomethylated genes. Sex-based analysis revealed hypomethylation in males at sites enriched in immune response pathways, with over 80% of differentially methylated positions in male individuals with childhood-onset lupus validated in the replication cohort.
Conclusion: Childhood-onset lupus is marked by significant hypomethylation in interferon-regulated genes, with DNA methylation changes correlating with disease activity and revealing distinct epigenetic molecular subgroups. Sex-specific patterns further suggest epigenetic contributions to sex-based disease heterogeneity in lupus.
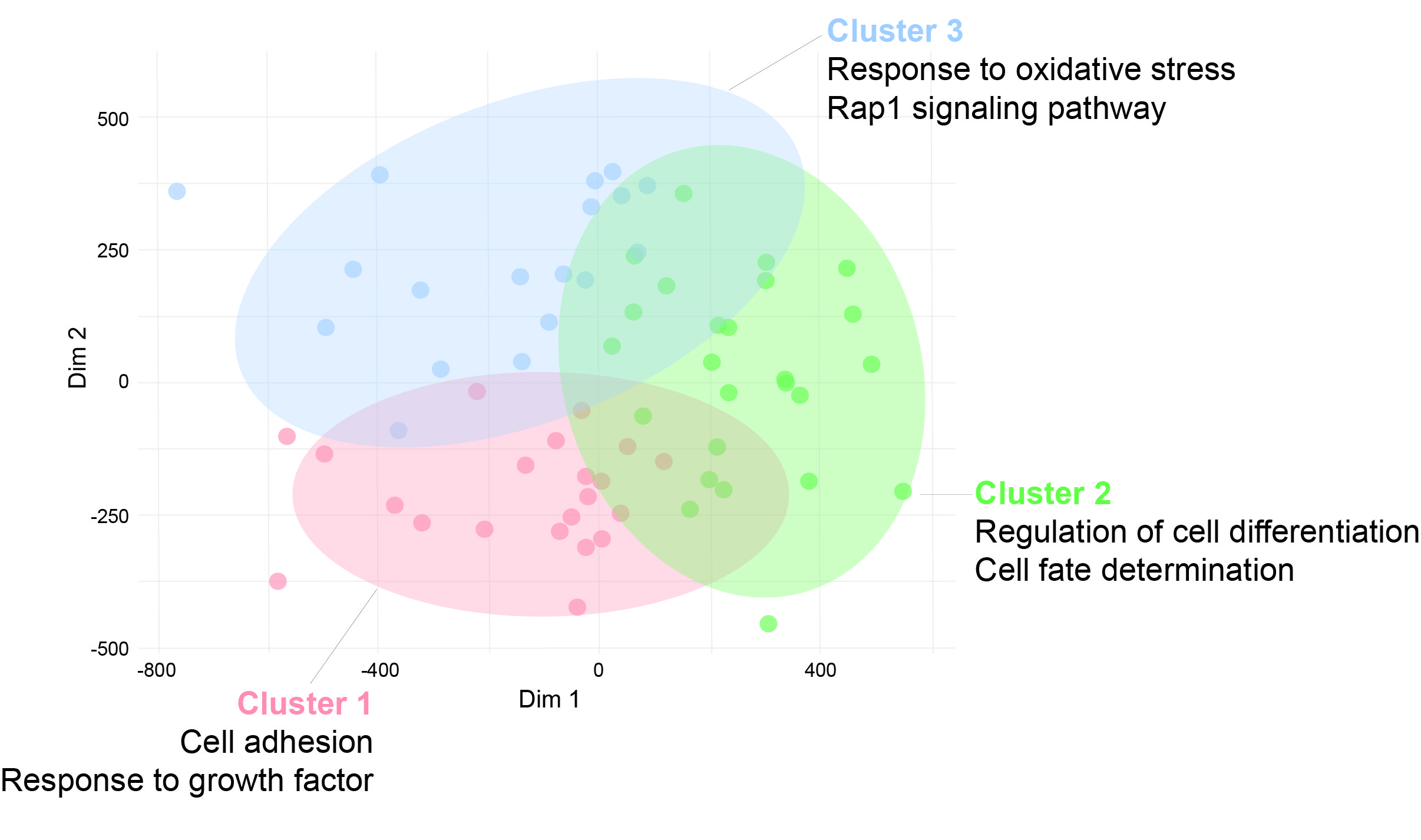 K-means clustering of patients with childhood-onset lupus based on DNA methylation profiles. Hypomethylated CpG sites characteristic of each cluster were analyzed for functional enrichment, revealing significant overrepresentation in Gene Ontology (GO) biological processes and KEGG pathways.
K-means clustering of patients with childhood-onset lupus based on DNA methylation profiles. Hypomethylated CpG sites characteristic of each cluster were analyzed for functional enrichment, revealing significant overrepresentation in Gene Ontology (GO) biological processes and KEGG pathways.
To cite this abstract in AMA style:
Casares Marfil D, Kavrul Kayaalp G, Guliyeva V, Akgün Ö, Türkmen Ş, Kilic Konte E, Şener S, Sahin S, Kasapcopur O, Sozeri B, Sözer Tokdemir S, Özen S, Aktay Ayaz N, Sawalha A. Epigenetic Profiling of Childhood-onset Lupus Reveals Distinct Epigenetic Clusters and Suggests Epigenetic Drivers of Disease Activity [abstract]. Arthritis Rheumatol. 2025; 77 (suppl 9). https://acrabstracts.org/abstract/epigenetic-profiling-of-childhood-onset-lupus-reveals-distinct-epigenetic-clusters-and-suggests-epigenetic-drivers-of-disease-activity/. Accessed .« Back to ACR Convergence 2025
ACR Meeting Abstracts - https://acrabstracts.org/abstract/epigenetic-profiling-of-childhood-onset-lupus-reveals-distinct-epigenetic-clusters-and-suggests-epigenetic-drivers-of-disease-activity/
