Session Information
Session Type: Poster Session B
Session Time: 10:30AM-12:30PM
Background/Purpose: Validated rheumatoid arthritis (RA) disease activity measures such as the Disease Activity Score-28 (DAS28) are used in clinical trials but not universally recorded in routine clinical care. Lack of readily available disease activity measures limits the use of electronic health records (EHRs) for generating real-world evidence (RWE) on RA treatment effectiveness. However, the key components of RA disease activity measures such as DAS28, including physical examination findings and laboratory assessments, are often documented in the EHRs, presenting an opportunity to determine disease activity using computational algorithms. The objective of this study was to produce scalable EHR RA disease activity metrics and conduct a test case comparing outcomes among RA patients switching from one tumor necrosis factor inhibitor (TNFi) to another vs changing mechanism of action (MOA).
Methods: We used data from a large EHR-based RA cohort (n=35,235) from a large multihospital healthcare system in the United States with natural language processing of clinical notes, coded EHR data, and laboratory results. A subset of these patients (n=1,579) was co-enrolled in an established RA disease registry and had planned assessment of RA disease activity by formal DAS28-CRP every 6 months. We developed a scalable machine learning imputed disease activity score (miDAS) using EHRs, computed aggregating semi-supervised and supervised learning approaches that combine DAS28-CPR measurements from the registry subset and large RA EHR cohort without DAS28-CRP. Using miDAS, we defined longitudinal moderate to severe RA disease activity (DAS28-CRP >3.2 or miDAS >0.365) over the full EHR cohort. We identified the patients who initiated TNFi and either 1) cycled to a second TNFi or 2) switched to a biologic DMARD or targeted-synthetic DMARD with another MOA. We then conducted the RWE study comparing the disease activity following the two treatment strategies. The primary outcome was the rate of medium to high disease activity 1 year after treatment switching adjusted for potential confounding using doubly robust causal modeling.
Results: A total of 2,119 RA patients were included with baseline characteristics listed in Table 1. Cross-validated against registry outcomes, miDAS achieved area-under-reception-operator-curve (AUC) for classifying moderate to high disease activity 0.829 (Table 2). There was no significant difference in risk of moderate to high disease activity at 1 year between TNFi cycling vs. MOA switching (average risk difference 1.9% [95% CI, -1.2% to 5.8%]) (Figure 1).
Conclusion: We used longitudinal electronic health records data and computational algorithms to derive and validate a disease activity measure (miDAS) with strong agreement with DAS28-CRP scores recorded in registry. This technique will allow for future real-world studies assessing RA treatments with imputed disease activity as an outcome. We also performed a proof-of-concept study demonstrating that switching mechanism of action after discontinuation of first TNFi did not result in significant difference in 1-year disease activity compared to cycling to a second TNFi.
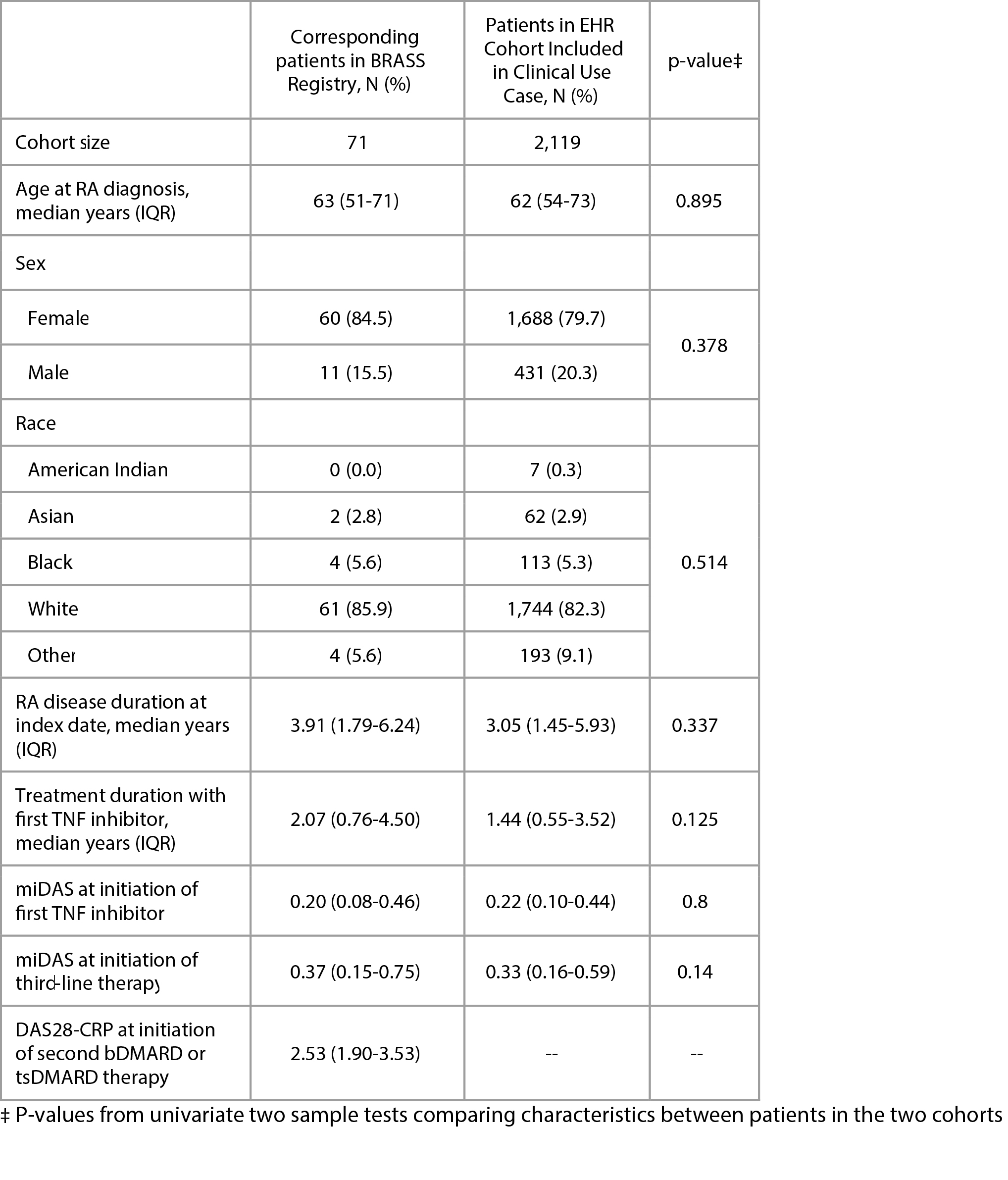 Patient characteristics according to data source
Patient characteristics according to data source
.jpg) Accuracy of miDAS and its machine learning components for classifying moderate to high disease activity as measured by area under curve
Accuracy of miDAS and its machine learning components for classifying moderate to high disease activity as measured by area under curve
.jpg) Treatment outcomes in clinical use case
Treatment outcomes in clinical use case
To cite this abstract in AMA style:
Hou J, Huang F, McDermott G, Wen J, Jeffway M, Qi Y, Han Y, Cai T, Bourgeois F, Liao K, Cai T. Measuring Rheumatoid Arthritis Outcomes Using Machine Learning Imputed Disease Activity Scores [abstract]. Arthritis Rheumatol. 2025; 77 (suppl 9). https://acrabstracts.org/abstract/measuring-rheumatoid-arthritis-outcomes-using-machine-learning-imputed-disease-activity-scores/. Accessed .« Back to ACR Convergence 2025
ACR Meeting Abstracts - https://acrabstracts.org/abstract/measuring-rheumatoid-arthritis-outcomes-using-machine-learning-imputed-disease-activity-scores/
