Session Information
Date: Monday, October 27, 2025
Title: (1306–1346) Rheumatoid Arthritis – Diagnosis, Manifestations, and Outcomes Poster II
Session Type: Poster Session B
Session Time: 10:30AM-12:30PM
Background/Purpose: Multiple Single-Nucleotide Polymorphisms (SNPs), including those in the methotrexate and azathioprine metabolic pathways, are associated with poor response to conventional disease-modifying anti-rheumatic drugs (cDMARDs) in Caucasians with rheumatoid arthritis. This retrospective cohort study aimed to investigate the association between SNPs and cDMARDs in responders and poor responders.
Methods: cDMARDs responders were defined as patients who had received standard treatment for at least 6 months and had not yet used biological agents. Poor responders were defined as patients who had received cDMARD therapy for at least 6 months and subsequently required treatment with biological agents. Genotype analysis was performed using Axiom Genome-Wide TPM 2.0 Array Plate, specifically designed for our population and contained 752,922 probes per array plat. A total of 38 SNPs were selected in AMPD, ATIC, DHFR, FPGS, GGH, ITPA, MTHFR, SLC19A1 and TYMS genes, which affect the methotrexate and azathioprine metabolic pathways. The distribution of allele frequency, odds ratio (OR), and 95% confidence intervals (CI) were calculated to evaluate the association between the categorical variables. A p-value of < 0.05 was considered to be statistically significant.
Results: Approximately 724,985.3 ± 416.1 (mean ± standard deviation) of SNP per sample met quality control requirements. SNPs affecting the methotrexate and azathioprine metabolic pathways were then selected. A total of 89 study participants including 66 DMARDs poor responders and 23 DMARDs responders were included in this study. There were no significant differences in age and sex between the two groups. Serum anti-CCP levels were significantly higher in the cDMARDs poor responder group (189.2 versus 103.7 U/mL, p = 0.011*). The distribution of allele frequency of the selected 38 SNPs and 2 in GHH (rs1800909 and rs3758149) were statistically different between the two groups (p =0.04062*, OR = 2.31, 95% CI: 0.91–5.31), and were associated with lower methotrexate efficacy.
Conclusion: The pharmacogenetics effects of cDMARDs poor responders could not be demonstrated in this study due to the small sample size. However, two SNPs (rs1800909 and rs3758149) in GHH which affect the methotrexate metabolic pathway were significantly different between the two groups. These SNPs exhibited a tendency towards an increasing OR. These genotypes can be presented as cDMARDs poor responders in our cohort.
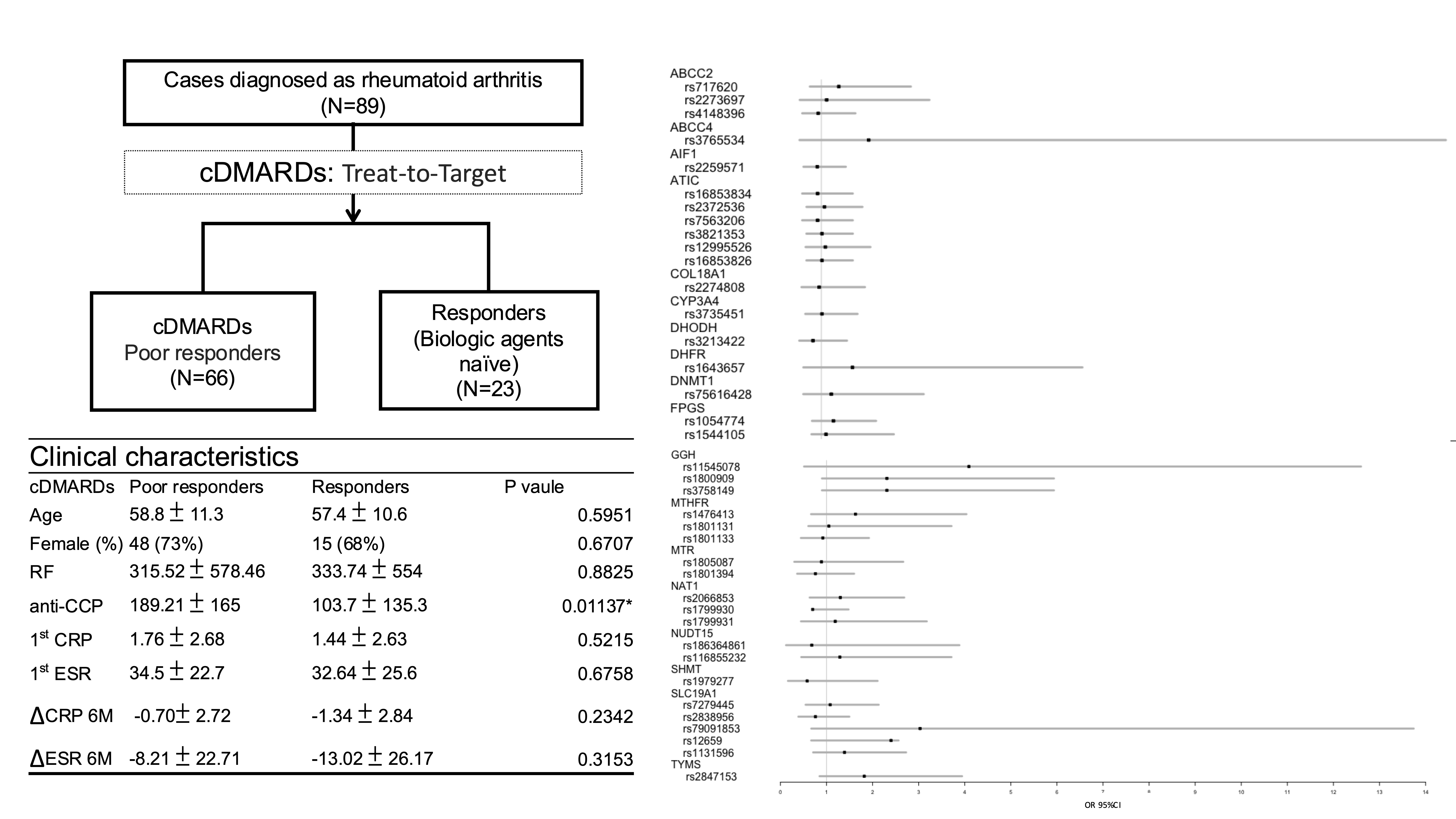 Flow chart and Result of OR CI95% illustrated by forest plot
Flow chart and Result of OR CI95% illustrated by forest plot
To cite this abstract in AMA style:
Chen Y, Wu C, Hsu J, Chen P. Preliminary Result of Investigating the Response of Conventional Disease-Modifying Antirheumatic Drugs (cDMARDs) Associated SNPs: One Cohort Study [abstract]. Arthritis Rheumatol. 2025; 77 (suppl 9). https://acrabstracts.org/abstract/preliminary-result-of-investigating-the-response-of-conventional-disease-modifying-antirheumatic-drugs-cdmards-associated-snps-one-cohort-study/. Accessed .« Back to ACR Convergence 2025
ACR Meeting Abstracts - https://acrabstracts.org/abstract/preliminary-result-of-investigating-the-response-of-conventional-disease-modifying-antirheumatic-drugs-cdmards-associated-snps-one-cohort-study/
