Session Information
Date: Monday, October 27, 2025
Title: (1306–1346) Rheumatoid Arthritis – Diagnosis, Manifestations, and Outcomes Poster II
Session Type: Poster Session B
Session Time: 10:30AM-12:30PM
Background/Purpose: Microbiome and microbial products are possible contributors and predictors to methotrexate (MTX) response variability. Bacterial small RNAs (sRNAs) are found in human plasma and can alter host gene expression and immune function. The purpose of this study was to determine 1) if circulating bacterial sRNAs are associated with response to MTX in patients with new onset RA (NORA), 2) whether sRNAs aligning to stool microbiome taxa that are associated with MTX response also are associated with MTX response, and 3) the overall relationship between circulating bacterial sRNAs and stool microbiome.
Methods: The study included a training cohort of 26 NORA and validation cohort of 20 NORA patients who were studied previously for relationship between microbiome and MTX response. Participants met ACR/EULAR 2010 classification criteria for RA, were treatment naïve at the time of plasma collection, and were followed for treatment response to MTX at 4 months. Plasma sRNAs from both cohorts (n=46) were sequenced and aligned to bacterial genomes using the TIGER pipeline. Individual bacterial sRNA sequences and taxa counts were compared in methotrexate responders (MTX-R) versus non-responders (MTX-NR) by DESeq2. Spearman’s rank correlation test was used to measure the association between plasma small RNAs and the stool microbiome.
Results: Demographic data are included in Table 1. Twelve bacterial sRNAs were enriched in the plasma of MTX-R versus MTX-NR in both the training and validation cohorts and included tRNA derived RNAs (tDRs) and ribosomal derived RNAs (rDRs) mapping to genera such as Prevotella, Bacteroides, Porphyromonas, Ruminococcus, Blautia, and Clostridium. Stool OTUs corresponding to Bacteroides and Prevotella, were abundant and significantly enriched in MTX-R versus MTX-NR in the training cohort as previously reported, and plasma sRNAs aligning to these taxa were also enriched in MTX-R versus MTX-NR in the same participants (p=0.04 and p=0.01, respectively). However, Clostridiales family and Ruminococcus genus that were decreased in MTX-R versus MTX-NR in the prior report were altered in plasma bacterial sRNAs of the same taxa but in the opposite direction (Figure 1). Combining training and validation cohorts, the overall correlation between the stool microbiome and respective plasma bacterial sRNA taxa counts was poor; however, several stool taxa were associated with the total abundance of bacterial sRNAs in plasma (Figure 2).
Conclusion: Consistent with findings that stool Prevotella and Bacteroides were increased in MTX-R, plasma sRNAs aligning to these taxa were similarly enriched in MTX-R versus MTX-NR. Moreover, increased abundance of specific plasma sRNAs aligning to Prevotella and Bacteroides were reproducibly enriched in MTX-R in two cohorts, positioning them as potential predictors of clinical response to MTX. Future studies will be needed to replicate these findings and determine the role of these specific bacterial sRNAs on MTX response and how they may gain entry to human circulation.
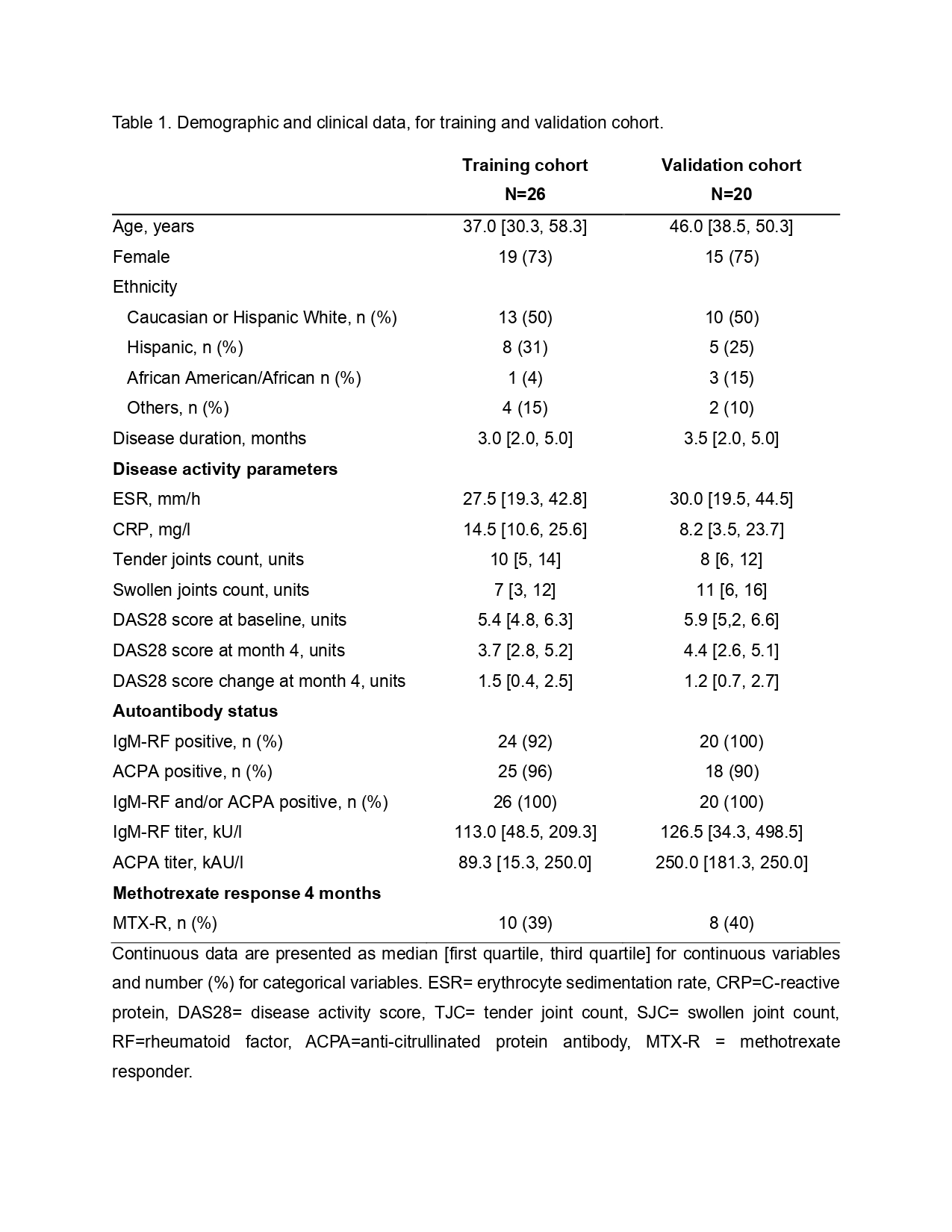 Table 1. Demographic and clinical data, for training and validation cohort
Table 1. Demographic and clinical data, for training and validation cohort
.jpg) Figure 1. Highly abundant (>0.01% in ≥1 group) and significantly altered stool OTUs in methotrexate responders (MTX-R) versus methotrexate non-responders (MTX-NR) (q < 0.05 by DESeq2) (as previously presented in Artacho et al Arthritis Rheumatol 2021;73(6):9310942) and their aligned plasma sRNAs in the training cohort.
Figure 1. Highly abundant (>0.01% in ≥1 group) and significantly altered stool OTUs in methotrexate responders (MTX-R) versus methotrexate non-responders (MTX-NR) (q < 0.05 by DESeq2) (as previously presented in Artacho et al Arthritis Rheumatol 2021;73(6):9310942) and their aligned plasma sRNAs in the training cohort.
.jpg) Stool microbiome genera and respective phylogeny significantly associated with the total abundance of bacterial sRNAs in plasma. Color scale refers to Spearman correlation coefficient.
Stool microbiome genera and respective phylogeny significantly associated with the total abundance of bacterial sRNAs in plasma. Color scale refers to Spearman correlation coefficient.
To cite this abstract in AMA style:
Ramirez-Becerra C, Joishy T, Chen S, Ramirez-Solano M, Sheng Q, Turnbaugh P, Ubeda C, Nayak R, Blank R, Scher J, Ormseth M. Relationship Between Circulating Bacterial Small RNAs, Methotrexate Response, and Microbiome in New Onset Rheumatoid Arthritis [abstract]. Arthritis Rheumatol. 2025; 77 (suppl 9). https://acrabstracts.org/abstract/relationship-between-circulating-bacterial-small-rnas-methotrexate-response-and-microbiome-in-new-onset-rheumatoid-arthritis/. Accessed .« Back to ACR Convergence 2025
ACR Meeting Abstracts - https://acrabstracts.org/abstract/relationship-between-circulating-bacterial-small-rnas-methotrexate-response-and-microbiome-in-new-onset-rheumatoid-arthritis/
