Session Information
Date: Monday, October 27, 2025
Title: (0955–0977) Systemic Sclerosis & Related Disorders – Basic Science Poster I
Session Type: Poster Session B
Session Time: 10:30AM-12:30PM
Background/Purpose: The poor prognosis of SSc patients demands an urgent need to prevent disease onset. Recently, we reported a global pro-inflammatory gene signature of dermal fibroblasts in pre-stages of SSc (pre-SSc). Here, we provide a detailed analysis of which fibroblast subtype and which molecular events are characteristic of this pro-inflammatory gene signature in pre-SSc.
Methods: We recruited patients with pre-SSc (n=6), established SSc (eSSc, n=7), and healthy controls (HC, n=6) (Table 1). Pre-SSc was defined according to the VEDOSS criteria1. eSSc patients met the 2013 ACR/EULAR criteria with disease duration < 5 years since their first non-Raynaud manifestation. HCs were matched for age, sex, and ethnicity. Site-matched forearm skin biopsies were collected, processed following the Chromium Next GEM protocol, and analyzed with Seurat pipeline. We refined fibroblast subclusters with functionally meaningful annotations. Over-Representation Analysis (ORA) of marker genes confirmed subcluster identities. Group-wise compositional differences were assessed (Speckle). Pairwise differential gene expression analysis was conducted (MAST, FDR < 0.05). Pathway analyses, ORA and Gene Set Enrichment Analysis (GSEA), were performed (clusterProfiler). Module scores (MS) quantified pro-inflammatory and pro-fibrotic gene expression programs.1. Avouac, Jérôme, et al. (2011), Annals of the rheumatic diseases.
Results: We identified 8 subtypes of dermal fibroblasts including type 1 (PF1), and type 2 (PF2) pro-inflammatory fibroblasts (Fig. 1A-C). Cell compositional analysis showed an increase in PF1 fibroblasts in pre-SSc (Fig. 1D). ORA of top marker genes confirmed functional differences among fibroblast subtypes and indicated a specific role of PF1 in cytokine-related immune response (Fig. 1E). In pre-SSc vs. HC, PF1 yielded 283 significant differentially expressed genes, including HLA-DRA, CD74, HLA-DPA1, HLA-DMA, and CCL21, consistent with findings in the global fibroblast analysis (Fig. 2A). Notably, HLA-DRA and HLA-DMA were also highly upregulated in eSSc vs. HC (Fig. 2B). Pre-SSc PF1 showed significantly higher expression of inflammatory response-related genes compared to HC and eSSc (Fig. 2C). In PF1, pre-SSc showed higher expression of pro-inflammatory genes vs. HC, enriched in 17/20 top pathways linked to cytokine/chemokine signaling, leukocyte and T cell activation, and antigen presentation via MHC complexes in ORA (Fig. 2D), further supported by GSEA results (Fig. 2E). Notably, similar signatures were also observed in eSSc vs. HC in ORA (Fig. 2F–G). MS gradients in pre-SSc and eSSc suggest a shared inflammatory mechanism, strongest in pre-SSc and shifting toward a pro-fibrotic profile in eSSc (Fig. 2H–I).
Conclusion: Pre-SSc fibroblasts show marked upregulation of pro-inflammatory gene signatures, primarily driven by the type 1 subtype. These signatures peak in pre-SSc before fibrosis and persist into early fibrotic SSc, highlighting a critical window for preventive intervention. The enrichment of immune response-related pathways in pre-SSc offers key insights into early SSc pathogenesis and the potential targets for preventive therapies.
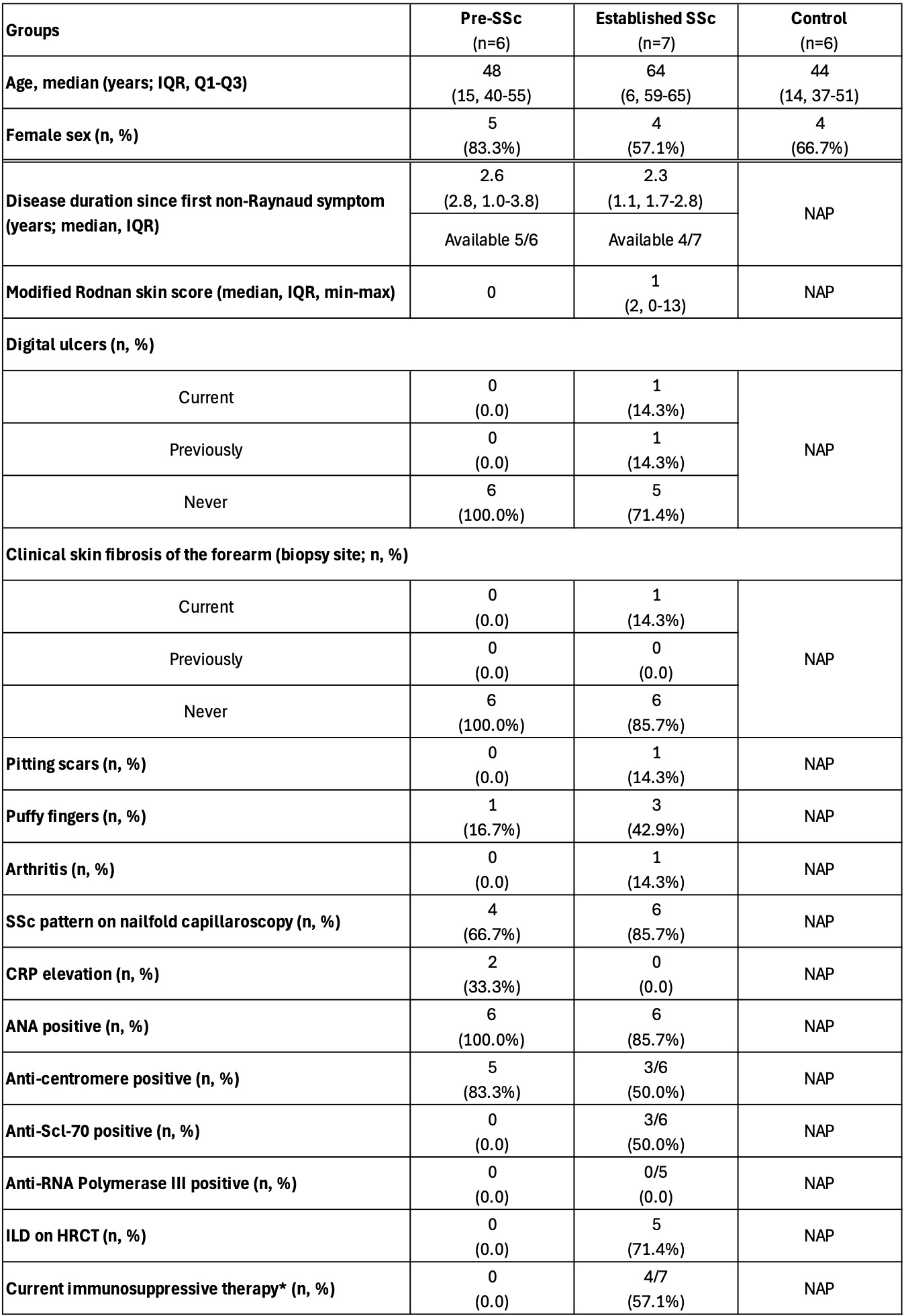 Table 1. Clinical characteristics of pre-SSc and eSSc patients, along with HCs, summarized by group. NAP: not applicable for HCs, indicating no assessment was performed in the healthy state; ILD: interstitial lung disease; HRCT: high-resolution computed tomography; CRP: C-reactive protein; ESR: erythrocyte sedimentation rate. *Immunosuppressive therapies included mycophenolate mofetil, tocilizumab, methotrexate, prednisone, or their combinations.
Table 1. Clinical characteristics of pre-SSc and eSSc patients, along with HCs, summarized by group. NAP: not applicable for HCs, indicating no assessment was performed in the healthy state; ILD: interstitial lung disease; HRCT: high-resolution computed tomography; CRP: C-reactive protein; ESR: erythrocyte sedimentation rate. *Immunosuppressive therapies included mycophenolate mofetil, tocilizumab, methotrexate, prednisone, or their combinations.
.jpg) Figure 1. A) UMAP showing 13 identified dermal cell types. B) UMAP of 8 identified fibroblast subtypes. C) Heatmap of the top 5 marker genes per subtype. Color bars correspond to the fibroblast subtypes shown in (B). D) Box plot showing fibroblast subtype proportions per patient across healthy controls (HC), pre-SSc, and early SSc (eSSc) groups. A trend toward increased PF1 fibroblasts was observed in pre-SSc compared to HC (proportional ratio: 1.37; p = 0.06; FDR = 0.3) E) GO over-representation analysis reveals distinct functions for the 8 fibroblast subtypes. The top 5 significant pathways per subtype are shown, based on marker genes identified by FindAllMarkers (default settings; Benjamini–Hochberg adjusted FDR < 0.05; average log2FC > 0.5).
Figure 1. A) UMAP showing 13 identified dermal cell types. B) UMAP of 8 identified fibroblast subtypes. C) Heatmap of the top 5 marker genes per subtype. Color bars correspond to the fibroblast subtypes shown in (B). D) Box plot showing fibroblast subtype proportions per patient across healthy controls (HC), pre-SSc, and early SSc (eSSc) groups. A trend toward increased PF1 fibroblasts was observed in pre-SSc compared to HC (proportional ratio: 1.37; p = 0.06; FDR = 0.3) E) GO over-representation analysis reveals distinct functions for the 8 fibroblast subtypes. The top 5 significant pathways per subtype are shown, based on marker genes identified by FindAllMarkers (default settings; Benjamini–Hochberg adjusted FDR < 0.05; average log2FC > 0.5).
.jpg) Figure 2. A) Volcano plot of differentially expressed genes (DEGs) in pre-SSc vs. healthy controls (HC), identified using the MAST model. Significant DEGs (|avg_log2FC| > 0.5; FDR < 0.05) are color-coded (blue: downregulated; orange: upregulated). Labeled genes meet stricter thresholds (|average log₂FC| ≥ 2; FDR < 0.0005). B) Volcano plot of DEGs in eSSc vs. HC, as described in (A). C) Expression levels of inflammatory response-related genes in type 1 pro-inflammatory fibroblasts across HC, pre-SSc, and eSSc. Genes shown are from the intersection of significant DEGs in global fibroblasts and type 1 fibroblasts in the pre-SSc vs. HC comparison. D–E) GO-based over-representation analysis (ORA) and gene set enrichment analysis (GSEA) of differentially expressed genes (DEGs) in type 1 fibroblasts (pre-SSc vs. HC). ORA was performed using DEGs with |avg_log₂FC| > 0.5 and FDR < 0.05. GSEA was based on DEGs ranked by score = avg_log₂FC + (−log₁₀FDR). FDR values above each NES bar indicate enrichment significance. F–G) ORA and GSEA of DEGs in type 1 fibroblasts (eSSc vs. HC), using the same thresholds and ranking method. H–I) Module scores across groups for pro-inflammatory (H) and pro-fibrotic (I) gene sets within type 1 fibroblasts. Pro-inflammatory genes include HLA-DRA, CD74, CXCL9, CXCL10, GBP1, HLA-DPA1, HLA-DRB1, PLAU, CCL21, TRAF1, GBP5, SLAMF8, CSF3, IFI30, OAS2, TLR2, IFIT2, IL4I1, CXCL11, HLA-DQA1; Pro-fibrotic genes include TNC, CTHRC1, COMP, COL4A4, CEMIP, LAMC2, WNT4, DDR1, COL4A3, IGFBP2, ADAM12, EDN1, IL23A, NOTCH4, CD37, PPARGC1A. **** indicates p < 0.0001.
Figure 2. A) Volcano plot of differentially expressed genes (DEGs) in pre-SSc vs. healthy controls (HC), identified using the MAST model. Significant DEGs (|avg_log2FC| > 0.5; FDR < 0.05) are color-coded (blue: downregulated; orange: upregulated). Labeled genes meet stricter thresholds (|average log₂FC| ≥ 2; FDR < 0.0005). B) Volcano plot of DEGs in eSSc vs. HC, as described in (A). C) Expression levels of inflammatory response-related genes in type 1 pro-inflammatory fibroblasts across HC, pre-SSc, and eSSc. Genes shown are from the intersection of significant DEGs in global fibroblasts and type 1 fibroblasts in the pre-SSc vs. HC comparison. D–E) GO-based over-representation analysis (ORA) and gene set enrichment analysis (GSEA) of differentially expressed genes (DEGs) in type 1 fibroblasts (pre-SSc vs. HC). ORA was performed using DEGs with |avg_log₂FC| > 0.5 and FDR < 0.05. GSEA was based on DEGs ranked by score = avg_log₂FC + (−log₁₀FDR). FDR values above each NES bar indicate enrichment significance. F–G) ORA and GSEA of DEGs in type 1 fibroblasts (eSSc vs. HC), using the same thresholds and ranking method. H–I) Module scores across groups for pro-inflammatory (H) and pro-fibrotic (I) gene sets within type 1 fibroblasts. Pro-inflammatory genes include HLA-DRA, CD74, CXCL9, CXCL10, GBP1, HLA-DPA1, HLA-DRB1, PLAU, CCL21, TRAF1, GBP5, SLAMF8, CSF3, IFI30, OAS2, TLR2, IFIT2, IL4I1, CXCL11, HLA-DQA1; Pro-fibrotic genes include TNC, CTHRC1, COMP, COL4A4, CEMIP, LAMC2, WNT4, DDR1, COL4A3, IGFBP2, ADAM12, EDN1, IL23A, NOTCH4, CD37, PPARGC1A. **** indicates p < 0.0001.
To cite this abstract in AMA style:
Li L, Pachera E, Dobrota R, Muraru S, Bürki K, Mihai C, Elhai M, Much L, Hofman A, Bearzi P, Hoffmann-Vold A, Distler O. Single-Cell RNA Sequencing Reveals a Prominent Pro-Inflammatory Gene Signature of Dermal Fibroblasts in Pre-Stages of SSc [abstract]. Arthritis Rheumatol. 2025; 77 (suppl 9). https://acrabstracts.org/abstract/single-cell-rna-sequencing-reveals-a-prominent-pro-inflammatory-gene-signature-of-dermal-fibroblasts-in-pre-stages-of-ssc/. Accessed .« Back to ACR Convergence 2025
ACR Meeting Abstracts - https://acrabstracts.org/abstract/single-cell-rna-sequencing-reveals-a-prominent-pro-inflammatory-gene-signature-of-dermal-fibroblasts-in-pre-stages-of-ssc/
