Session Information
Date: Sunday, October 26, 2025
Session Type: Abstract Session
Session Time: 3:45PM-4:00PM
Background/Purpose: The impact of genetic risk factors on rheumatoid arthritis (RA) phenotype is incompletely understood. Comparing individual genetic variants associated with RA susceptibility to a polygenic score (PGS), we hypothesized that higher genetic risk for RA would correlate with more severe disease.
Methods: We genotyped participants from the Veterans Affairs RA (VARA) registry, a prospective multicenter RA cohort. We evaluated the association between an existing non-HLA PGS (PGS Catalog PGS002745) and RA disease activity measured by the clinical disease activity index (CDAI), using linear regression adjusted for sex, age, and genetic background. We applied similar linear models to evaluate the association between individual genetic risk factors PTPN22 R620W or HLA-DRB1 shared epitope (SE) and disease activity scores. We tested the association with these genetic risk factors and binary outcomes (RF or anti-CCP seropositivity, erosive disease) at enrollment using logistic regression adjusted for the same covariates. To understand the potential for gene-phenotype interactions, we tested for an interaction of genetic exposure with a) disease duration and b) seropositivity (RF or anti-CCP). We also tested for interaction of SE with PTPN22 R620W and PGS.
Results: Among 2597 VARA participants, 50 (2%) were homozygous for PTPN22 R620W, and 1603 (62%) had a PGS >50th percentile compared to the 1000 Genomes reference population. PTPN22, SE, and the PGS were strongly associated with seropositivity (PGS OR= 1.41, 95% CI 1.26-1.58, p< 0.001, Fig. 1). Those with 2 PTPN22 alleles had higher disease activity (CDAI β 6.10, 95% CI 1.23-10.97, p=0.014); no difference was observed for those with one PTPN22 allele, SE, or by PGS (Fig. 2). Higher genetic risk appeared more strongly associated with disease severity in those with longer disease duration at enrollment. The association between 2 copies of PTPN22 and higher disease activity was only observed in those with >3 years disease duration (Fig. 3). The association between 2 PTPN22 alleles and higher disease activity was also primarily observed among seropositive patients (Fig. 3).The association between presence of 2 PTPN22 alleles and higher disease activity was stronger in the presence of SE (CDAI: effect with SE 9.39, 95% CI 3.75-15.0, p interaction 0.01, Fig. 3). No interactions were observed between seropositivity or SE and the PGS. The presence of SE was associated with erosive disease at study enrollment [1 allele: OR 1.40, 95% CI 1.13-1.74, p = 0.002; 2 allele OR 1.46, 95% CI 1.10-1.93, p = 0.009]; higher PGS and PTPN22 were not (Fig. 1). A higher PGS was associated with higher odds of erosive disease in those with >3 years duration (1 SD change OR 1.12, 95% CI 1.01-1.26, p = 0.035), but not among those with shorter disease duration.
Conclusion: Genetic risk factors for RA demonstrate heterogeneous associations with resultant disease phenotypes among RA patients. Genetic effects were most significant after the early RA period and among those who were seropositive. Individual genetic variants and pathway-specific genetic risk may be more informative than the genome-wide genetic risk for RA in understanding phenotype and disease activity in RA.
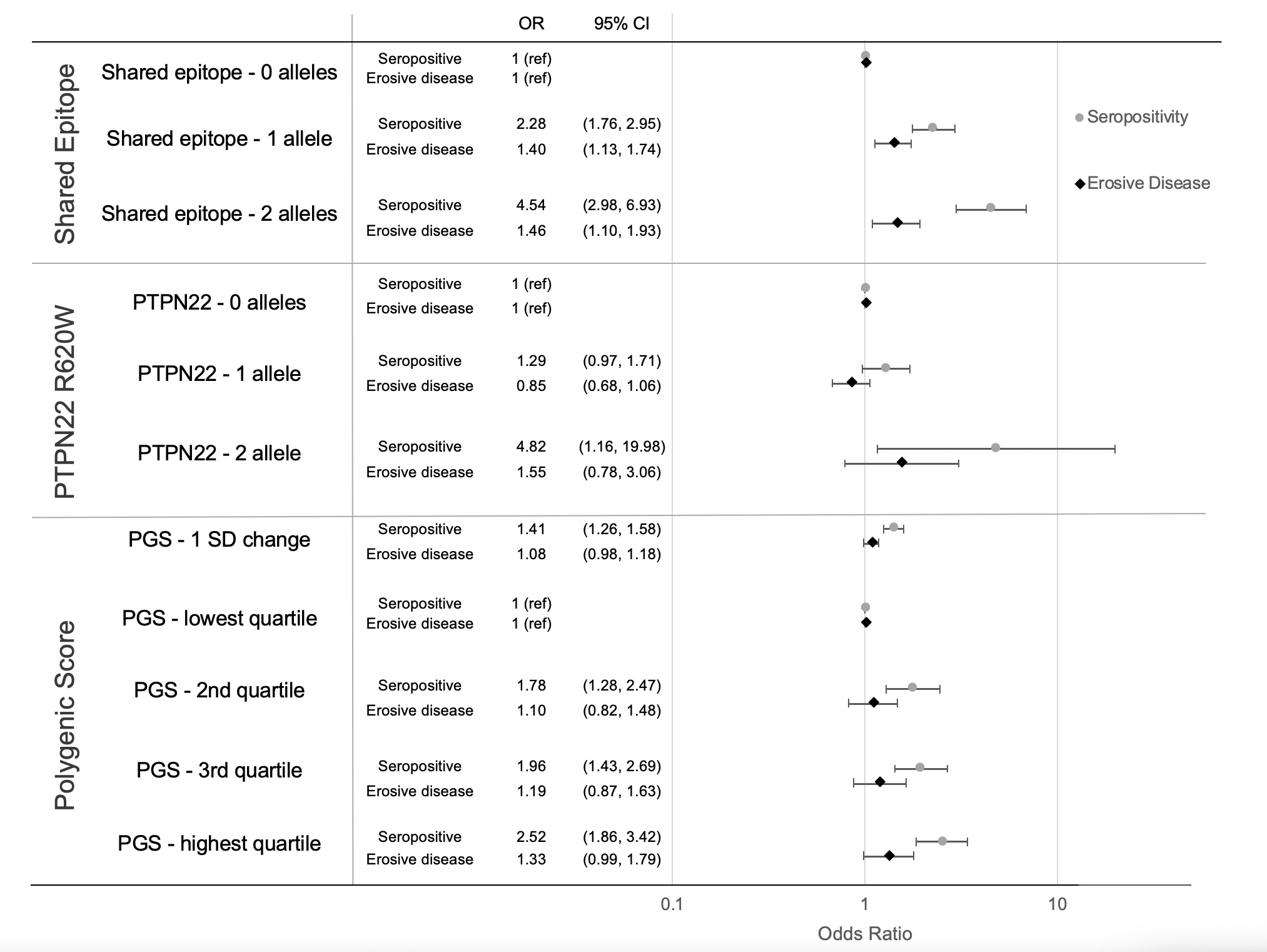 Figure 1. Effect of HLA-DRB1 SE, PTPN22 R620W, and non-HLA PGS on seropositivity and erosive disease at VARA enrollment.
Figure 1. Effect of HLA-DRB1 SE, PTPN22 R620W, and non-HLA PGS on seropositivity and erosive disease at VARA enrollment.
Effect determined by linear regression in a genotype model, adjusted for age, sex, and top 5 PCs of population structure. All genetic models demonstrated increased odds for seropositivity, whereas only SE was significantly associated with erosive disease.
Abbreviations:
PGS – polygenic score
SE – shared epitope
PC – principal components
OR – odds ratio
CI – confidence interval
.jpg) Figure 2. Effect of SE, PTPN22 R620W, and non-HLA PGS on CDAI at enrollment.
Figure 2. Effect of SE, PTPN22 R620W, and non-HLA PGS on CDAI at enrollment.
Participants with 2 copies of PTPN22 demonstrated a higher CDAI (p = 0.014). A trend toward a negative association with CDAI is observed for 1 standard deviation change of PGS ( p = 0.057) and in those at the highest quartile of risk by PGS) (p = 0.053). Effect at enrollment determined by linear regression in a genotype model, adjusted for age, sex, and top 5 PCs of population structure.
Abbreviations:
CDAI – clinical disease activity index
PGS – polygenic score
SE – shared epitope
PC – principal components
CI – confidence interval
.jpg) Figure 3. Subgroup Analysis for PTPN22 R620W and CDAI.
Figure 3. Subgroup Analysis for PTPN22 R620W and CDAI.
Subgroup analysis performed by stratification in linear regression in a genotype model, adjusted for age, sex, and top 5 PCs of population structure. Longitudinal disease activity was analyzed using generalized estimating equations adjusting for these same factors as well as enrollment CDAI. Participants with 2 copies of PTPN22 demonstrated a higher CDAI at enrollment, but not in longitudinal disease activity corrected for the enrollment CDAI. This effect was predominant in those with seropositive RA, those with 1 or 2 shared epitope HLA alleles, and with disease duration ≥ 3 years. There was not a significant effect of genotype in early disease. Those with 1 PTPN22 R620W allele with seronegative RA had significantly lower disease activity, and a trend toward lower disease activity in those who were seronegative or without shared epitope with 2 PTPN22 R620W alleles, though this was not statistically significant.
Abbreviations:
CDAI – clinical disease activity index
VARA – Veterans Affairs Rheumatoid Arthritis Registry
SE – shared epitope
PC – principal components
To cite this abstract in AMA style:
Riley T, Wheeler A, England B, Cannon G, Sauer B, Kunkel G, Wysham K, Wallace B, Monach P, Reimold A, Kerr G, Smith I, Richards J, Lee I, Thiele G, Xiao R, Damrauer S, Levin M, George M, Mikuls T, Baker J. Heterogeneity in the Association of Genetic Risk for Rheumatoid Arthritis and Resultant Rheumatoid Arthritis Phenotypes [abstract]. Arthritis Rheumatol. 2025; 77 (suppl 9). https://acrabstracts.org/abstract/heterogeneity-in-the-association-of-genetic-risk-for-rheumatoid-arthritis-and-resultant-rheumatoid-arthritis-phenotypes/. Accessed .« Back to ACR Convergence 2025
ACR Meeting Abstracts - https://acrabstracts.org/abstract/heterogeneity-in-the-association-of-genetic-risk-for-rheumatoid-arthritis-and-resultant-rheumatoid-arthritis-phenotypes/
