Session Information
Session Type: Poster Session A
Session Time: 10:30AM-12:30PM
Background/Purpose: Rheumatoid arthritis (RA) shows heterogeneous therapeutic responses that may reflect diversity within synovial tissue (ST). We aimed to identify gene signatures that predict Adalimumab response and provide an initial spatial transcriptomic validation of their cellular context.
Methods: ST biopsies were obtained from 30 anti-citrullinated protein antibody–positive, treatment-naïve RA patients (symptom duration ≤ 12 months) classified by immunohistochemistry as TNF- or T-cell-dominant. Bulk RNA-seq data for all protein-coding genes were analysed with glmnet LASSO-regularised logistic regression to derive a small set of genes predictive of remission versus non-remission using baseline samples from patients targeted for Adalimumab treatment (remission and non-remission at 3 months; DAS CRP< 2.6). The resulting model was also applied on baseline samples from Abatacept and triple-DMARD non-responders. The predictive genes were mapped onto high-resolution Stereo-seq spatial transcriptomic sections. Spatially resolved expression was assessed against cell-type annotations derived from unsupervised clustering and marker genes.
Results: A concise set of genes clearly separated early-remission from non-remission cases in the Adalimumab cohort. When applied to baseline samples from other treatment arms, the predictive model identified one patient in the Abatacept cohort and six patients in the triple-DMARD cohort as having remission-like profiles, indicating scope for treatment re-stratification. Preliminary Stereo-seq analysis suggests these genes may be more locally enriched in baseline ST from early-remission cases (Figure 1).
Conclusion: This study identifies key synovial gene signature that predicts clinical response to Adalimumab and demonstrate, for the first time, its restricted spatial expression in specific ST cell populations. Although preliminary, these Stereo-seq data strengthen the biological plausibility of the signature and support future validation in larger spatial cohorts. Incorporating qPCR-based testing of these genes in routine biopsies could expedite personalised therapeutic decisions in RA.
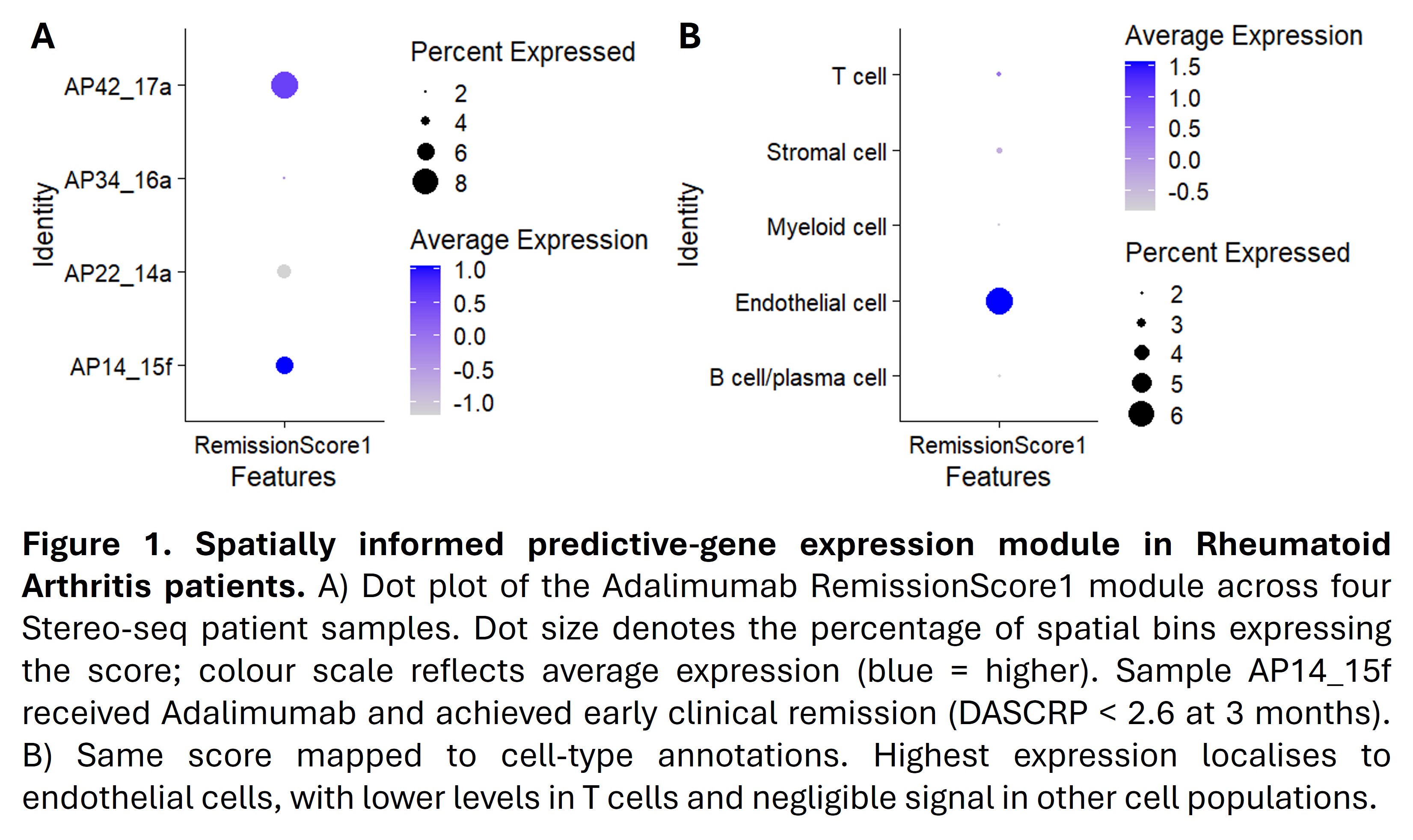 Spatially informed predictive‐gene expression module in Rheumatoid Arthritis patients.
Spatially informed predictive‐gene expression module in Rheumatoid Arthritis patients.
To cite this abstract in AMA style:
Wong V, Lowe K, Small A, Altmann C, Proudman S, Smith M, Suwakulsiri W, Thomas R, Kok C, Wechalekar M. Spatial profiling of gene signatures in synovial tissue informs treatment strategy for Rheumatoid Arthritis [abstract]. Arthritis Rheumatol. 2025; 77 (suppl 9). https://acrabstracts.org/abstract/spatial-profiling-of-gene-signatures-in-synovial-tissue-informs-treatment-strategy-for-rheumatoid-arthritis/. Accessed .« Back to ACR Convergence 2025
ACR Meeting Abstracts - https://acrabstracts.org/abstract/spatial-profiling-of-gene-signatures-in-synovial-tissue-informs-treatment-strategy-for-rheumatoid-arthritis/
