Session Information
Session Type: Poster Session A
Session Time: 10:30AM-12:30PM
Background/Purpose:
Compared to the low-risk genotype (LRG) the APOL1 high-risk genotype (HRG) confers an increased risk of end stage kidney disease among individuals with systemic lupus erythematosus (SLE). Despite this association, the cellular and molecular mechanisms driving APOL1-mediated cytotoxicity in SLE remain unclear. Monocytes both express APOL1 and are exposed to the SLE inflammatory milieu. This study aimed to define differential pathways of monocyte cytotoxicity in SLE patients stratified by APOL1 genotype.
Methods:
THP-1 monocytes were stimulated with IFN-γ, and APOL1 expression was quantified via qRT-PCR and Western blot. Monocytes from SLE patients were isolated using Ficoll density gradient and magnetic bead separation. Blood DNA was extracted from patients and genotyped for APOL1 risk variants. Type-I interferon activity in plasma was assessed using the WISH cell bioassay. RNA-seq was performed on patient’s monocytes, and differential gene expression analysis compared high-risk (2 APOL1 variants) and zero-risk groups, followed by gene ontology and pathway enrichment analyses.
Results: Demographic and clinical data from 23 SLE patients were stratified by APOL1 risk allele count (0, 1, or 2). Those with 2 risk variants (n=5) had lower mean eGFR (64 ± 28.5 mL/min/1.73 m²) and SLEDAI scores (4 ± 2) than patients with 0 (97 ± 15.6; 7.7 ± 5.7) or 1 variant (105.4 ± 18.2; 8.4 ± 6.3). Type I interferon activity, assessed via WISH cell assay, declined with increasing APOL1 variants (mean: 0RV 9.28, 1RV 4.25, 2RV 1.34; p=0.07). IFN-γ treatment of THP-1 monocytes confirmed APOL1 expression, showing 31.1-fold (qRT-PCR) and 2.5-fold (western blot) increases. Transcriptomic profiling revealed that monocytes from high-risk genotype (HRG) patients upregulated genes in TNF-α signaling, coagulation, inflammation, and cholesterol homeostasis. Key upregulated genes included CD5L, TRAP1, HLA-DQA, TNFRSF19, KIR3DL2, and platelet receptor, P2RY12. Conversely, HLA-DRB5, IFNGR2, and TNFSF13B (BAFF) were downregulated in HRG vs low-risk genotype (LRG) monocytes.
Conclusion: Our findings indicate that circulating monocytes from HRG SLE patients exhibit a transcriptomic profile characterized by proinflammatory and procoagulant signatures, alongside diminished antigen presentation capacity. These results provide novel insights into how APOL1 risk variants may contribute to cytotoxicity in SLE beyond classical interferon-driven mechanisms.
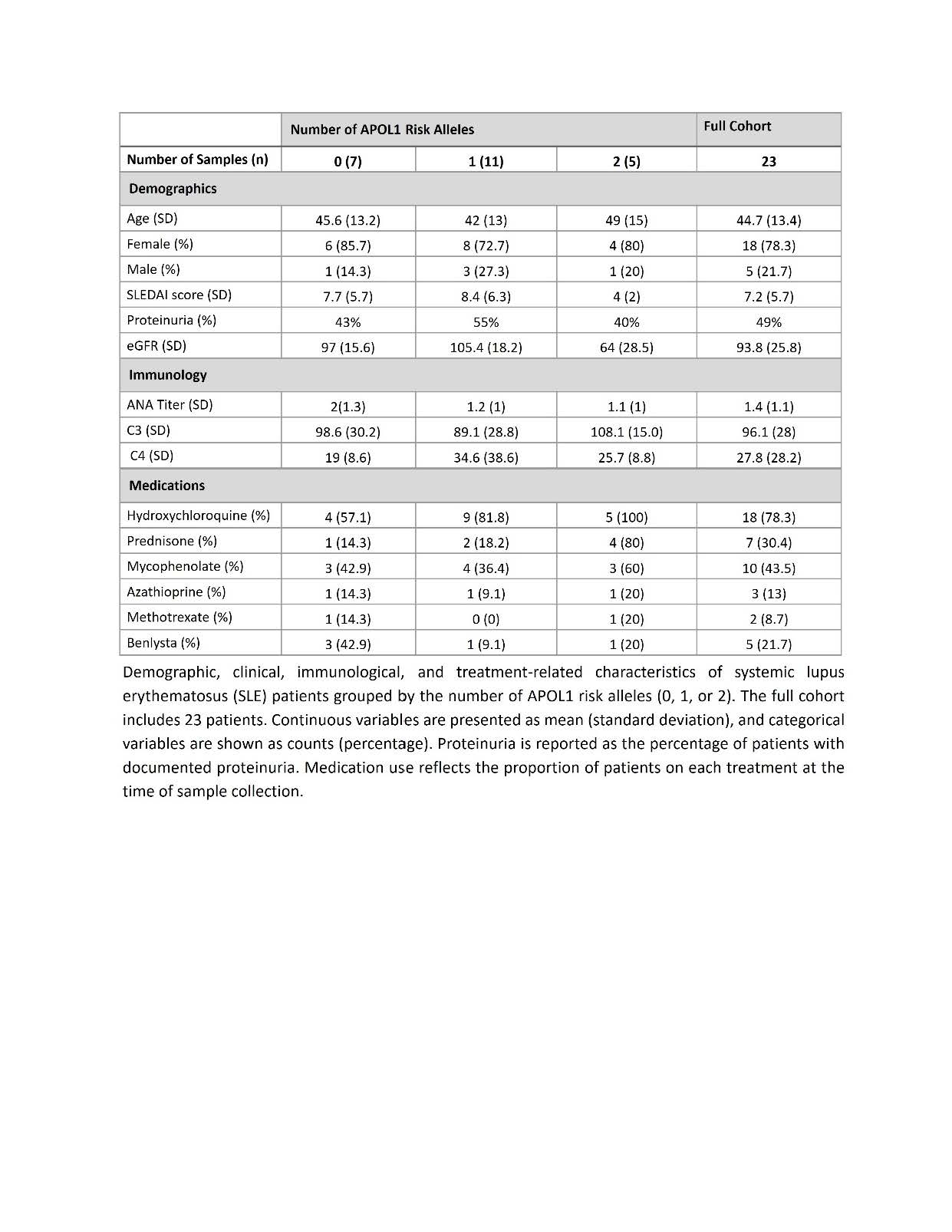 Table-1 Clinical and Demographic Characteristics of SLE Patients Stratified by APOL1 Risk Alleles
Table-1 Clinical and Demographic Characteristics of SLE Patients Stratified by APOL1 Risk Alleles
To cite this abstract in AMA style:
Pandian V, Agboola A, ramani S, Felix C, Nimoni A, Divers J, Niewold T, Blazer A. Monocyte Transcriptomic Signatures Uncover Potential Pathogenic Mechanisms of the APOL1 High Risk Genotype (HRG) [abstract]. Arthritis Rheumatol. 2025; 77 (suppl 9). https://acrabstracts.org/abstract/monocyte-transcriptomic-signatures-uncover-potential-pathogenic-mechanisms-of-the-apol1-high-risk-genotype-hrg/. Accessed .« Back to ACR Convergence 2025
ACR Meeting Abstracts - https://acrabstracts.org/abstract/monocyte-transcriptomic-signatures-uncover-potential-pathogenic-mechanisms-of-the-apol1-high-risk-genotype-hrg/

.jpg)