Session Information
Session Type: Poster Session A
Session Time: 10:30AM-12:30PM
Background/Purpose: Patients with rheumatic conditions are at increased risk for cardiovascular (CV) problems, striking on average a decade before peers and conferring substantial morbidity and mortality. Understanding the mechanisms underlying this elevated CV risk could lead to improved care for rheumatology patients and novel treatments for CV conditions. While genetic studies have illuminated several mutual risk loci between certain pairs of conditions, these studies have been limited in size, scope, and replication. Our goal was to conduct a comprehensive joint genetic analysis across 8 rheumatic conditions and 8 CV conditions based on two nation-wide data repositories, characterizing the shared genetic risk, loci, and putative causal SNPs.
Methods: We used the Million Veteran Program (MVP; n = 453,878) and UK Biobank (UKB; n = 406,492) EUR cohorts and genome-wide association study (GWAS) statistics based on PheCode definitions of 8 rheumatic (rheumatoid arthritis [RA], lupus [SLE], ankylosing spondylitis [AS], psoriatic arthropathy [PA], regional enteritis, sicca syndrome, ulcerative colitis [UC], and chronic UC) and 8 CV traits (coronary artery disease [CAD], arterial embolism, aortic aneurysm [AA], venous embolism, myocardial infarction [MI], congestive heart failure [CHF], peripheral vascular disease [PVD], and cerebrovascular disease [CVD]) (MVP: Verma et al. 2024, UKB: Zhou et al. 2018) (Figure 1). Genetic heritability (LDSC), global genetic correlation (LDSC), local genetic correlation (HESS), systematic colocalization (coloc, with adaptive locus identification), fine-mapping (SuSiE), and functional enrichment (STRING) were performed for each rheumatic-CV trait pair in a bidirectional replication framework.
Results: Heritabilities were generally higher in MVP than UKB, particularly for CV traits. A total of 31 significant genetic correlations (false discovery rate [FDR] < 0.05) were identified, all positive. Four rheumatic-CV trait pair genetic correlations replicated in the reverse cohort setup (all involving RA; range: 0.08 – 0.30). A total of 76 regions showed nominal local genetic correlation (P < 0.05), with 23 of those regions at the 6p21.32/6p21.33 locus. All genome-wide-significant SNPs (P < 5e-8) were tested for colocalization (posterior probability for H3 or H4 > 0.8) and fine-mapped (4,773 loci), yielding 25 loci estimated to share the same causal variant (H3) and 26 loci estimated to have different causal variants (H4). These 51 loci were functionally enriched for genes related to antigen processing and peptide presentation (Gene Ontology BP; FDR < 0.05). Implicated loci for RA included ATXN2 (with CAD, MI, CVD, venous embolism), SH2B3 (with MI, PVD), RSBN1 (with CHF), and the 11p34.1 locus near ST3GAL4 and KIRREL3 (with CAD).
Conclusion: Our findings support the presence of shared genetic risk between numerous rheumatic and CV conditions, both on the level of mutually implicated genes and of potentially causal variants. Genes related to antigen processing and peptide presentation were enriched among the colocalized loci of rheumatic and CV traits. The strongest shared genetic signal was seen for RA, implicating ATXN2, SH2B3, RSBN1, and the 11p34.1 locus.
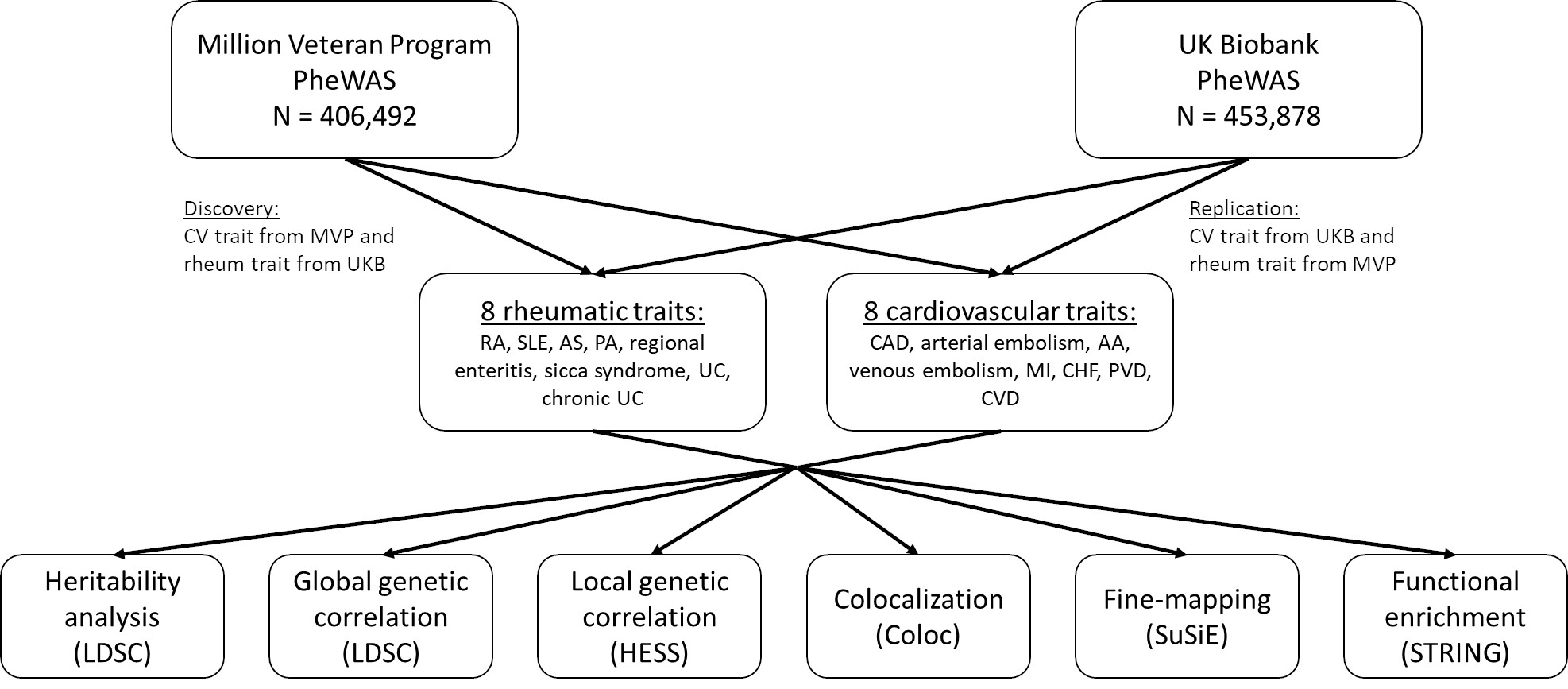 Figure 1. Study design. Genome-wide association study (GWAS) summary statistics from PheWAS performed in both MVP (Verma et al. 2024) and UKB (Zhou et al. 2018) for 16 total rheumatic and CV traits were used. The rheumatic traits included rheumatoid arthritis (RA), systemic lupus erythematosus (SLE), ankylosing spondylitis (AS), psoriatic arthropathy (PA), regional enteritis, sicca syndrome, ulcerative colitis (UC), and chronic UC. The CV traits included coronary artery disease (CAD), arterial embolism and thrombosis, aortic aneurysm, other venous embolism and thrombosis, myocardial infarction MI), congestive heart failure (nonhypertensive) (CHF), peripheral vascular disease (PVD), and cerebrovascular disease (CVD). For the discovery analysis, the CV trait was taken from MVP and the rheumatic trait taken from UKB; the replication analysis reversed this order.
Figure 1. Study design. Genome-wide association study (GWAS) summary statistics from PheWAS performed in both MVP (Verma et al. 2024) and UKB (Zhou et al. 2018) for 16 total rheumatic and CV traits were used. The rheumatic traits included rheumatoid arthritis (RA), systemic lupus erythematosus (SLE), ankylosing spondylitis (AS), psoriatic arthropathy (PA), regional enteritis, sicca syndrome, ulcerative colitis (UC), and chronic UC. The CV traits included coronary artery disease (CAD), arterial embolism and thrombosis, aortic aneurysm, other venous embolism and thrombosis, myocardial infarction MI), congestive heart failure (nonhypertensive) (CHF), peripheral vascular disease (PVD), and cerebrovascular disease (CVD). For the discovery analysis, the CV trait was taken from MVP and the rheumatic trait taken from UKB; the replication analysis reversed this order.
.jpg) Figure 2. Heritability and global genetic correlation between rheumatic and cardiovascular traits. (A) The heritabilities estimated by LD score regression (LDSC) in a EUR population for each trait (defined by PheCode) are shown by cohort. The pairwise global genetic correlations between each rheumatic and CV trait are shown for both the discovery (B) (CV trait from MVP and rheumatic trait from UKB) and replication (C) (CV trait from UKB and rheumatic trait from MVP) analyses. Statistically significant correlations (FDR < 0.05) are indicated by text showing the estimated correlation in the heatmap. Gray indicates failed estimations (likely due to low heritability in one or both traits).
Figure 2. Heritability and global genetic correlation between rheumatic and cardiovascular traits. (A) The heritabilities estimated by LD score regression (LDSC) in a EUR population for each trait (defined by PheCode) are shown by cohort. The pairwise global genetic correlations between each rheumatic and CV trait are shown for both the discovery (B) (CV trait from MVP and rheumatic trait from UKB) and replication (C) (CV trait from UKB and rheumatic trait from MVP) analyses. Statistically significant correlations (FDR < 0.05) are indicated by text showing the estimated correlation in the heatmap. Gray indicates failed estimations (likely due to low heritability in one or both traits).
.jpg) Figure 3. Colocalization and fine-mapping of genetic loci underlying rheumatic and cardiovascular traits. (A) The local genetic correlation for each tested region and for all 128 trait pairs is shown. The color indicates the rheumatic trait, and the shape indicates the CV trait. Significant enrichment of correlated loci with bidirectional replication was found in the 6p21.32/6p21.33 region. (B) The posterior probabilities of both traits being associated (y axis; sum of H3 and H4 coloc hypotheses) for each of the 4,773 tested loci are shown. For significantly colocalized loci (posterior probability > 0.8), the label shows the nearest gene. Insignificant loci are shown as gray points. (C) A functional gene network (STRING) of the genes implicated by the colocalization analysis is shown, including genes from loci either with shared or different casual variants (H3 or H4 > 0.8). (D) The GWAS assocations for the 11p34.1 locus underlying both RA and CAD are shown (results show discovery analysis). The red line indicates the genome-wide significance threshold (5e-8). The blue lines and labels indicate protein-coding genes near the locus (GENCODE).
Figure 3. Colocalization and fine-mapping of genetic loci underlying rheumatic and cardiovascular traits. (A) The local genetic correlation for each tested region and for all 128 trait pairs is shown. The color indicates the rheumatic trait, and the shape indicates the CV trait. Significant enrichment of correlated loci with bidirectional replication was found in the 6p21.32/6p21.33 region. (B) The posterior probabilities of both traits being associated (y axis; sum of H3 and H4 coloc hypotheses) for each of the 4,773 tested loci are shown. For significantly colocalized loci (posterior probability > 0.8), the label shows the nearest gene. Insignificant loci are shown as gray points. (C) A functional gene network (STRING) of the genes implicated by the colocalization analysis is shown, including genes from loci either with shared or different casual variants (H3 or H4 > 0.8). (D) The GWAS assocations for the 11p34.1 locus underlying both RA and CAD are shown (results show discovery analysis). The red line indicates the genome-wide significance threshold (5e-8). The blue lines and labels indicate protein-coding genes near the locus (GENCODE).
To cite this abstract in AMA style:
Panyard D, Li D, Kho P, Guarischi-Sousa R, Zhou J, Hilliard A, Bartels C, Tsao P, Assimes T. Biobank-scale genetic mapping identifies the shared genetic landscape of rheumatic and cardiovascular disease [abstract]. Arthritis Rheumatol. 2025; 77 (suppl 9). https://acrabstracts.org/abstract/biobank-scale-genetic-mapping-identifies-the-shared-genetic-landscape-of-rheumatic-and-cardiovascular-disease/. Accessed .« Back to ACR Convergence 2025
ACR Meeting Abstracts - https://acrabstracts.org/abstract/biobank-scale-genetic-mapping-identifies-the-shared-genetic-landscape-of-rheumatic-and-cardiovascular-disease/
