Session Information
Session Type: Poster Session C
Session Time: 10:30AM-12:30PM
Background/Purpose: Myositis is a term for a heterogeneous group of inflammatory myopathies, each with characteristic clinical manifestations, histopathological features, and autoantibodies, including myositis-specific autoantibodies (MSAs). Despite the increasing use of targeted therapeutics, few studies have comprehensively examined subtype-specific muscle expression of inflammatory mediators that could serve as targets for these agents. In this study, we perform a large RNA sequencing (RNA-seq) analysis to identify cytokines, cytokine receptors, and checkpoint genes specific to each myositis subtype.
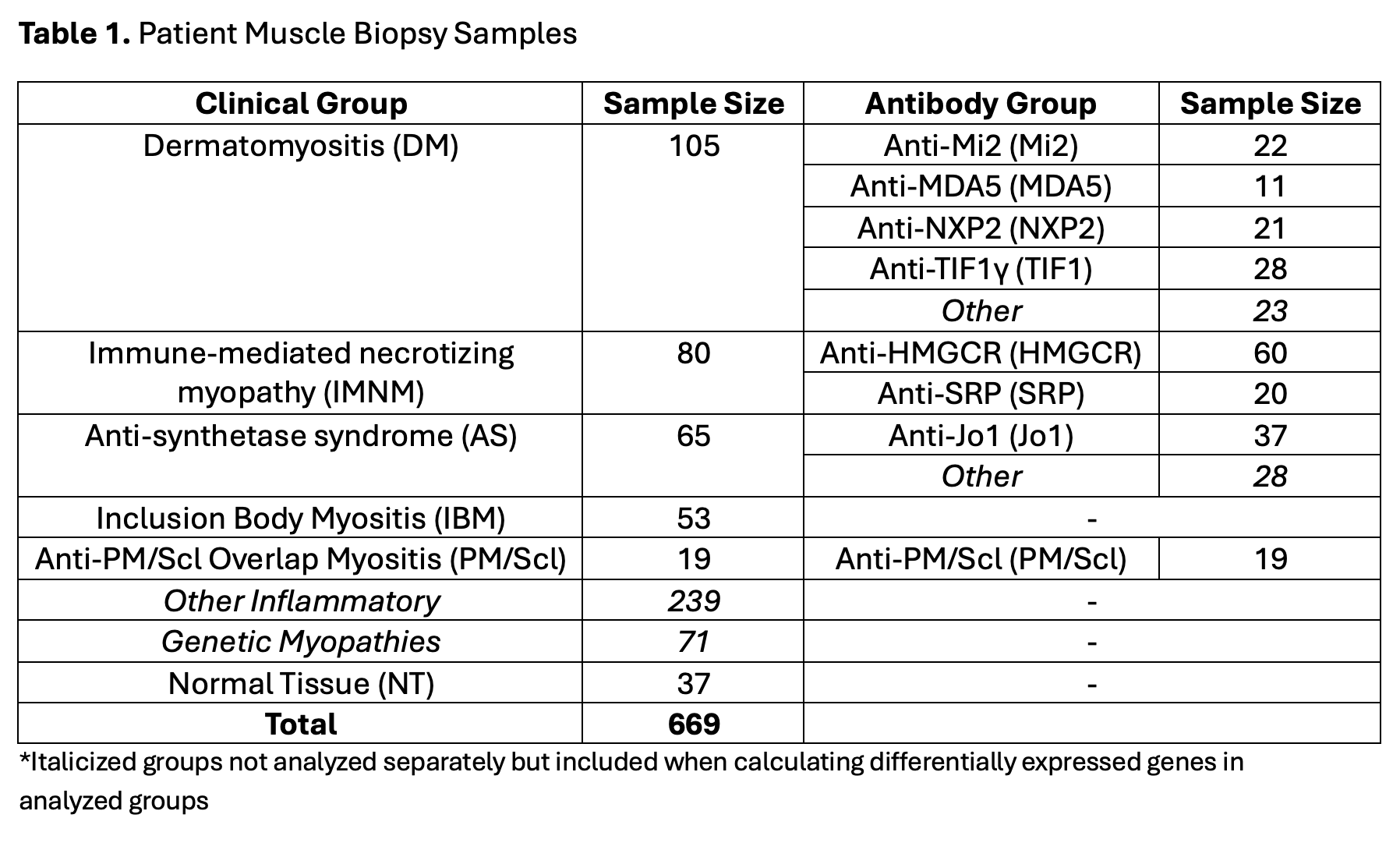
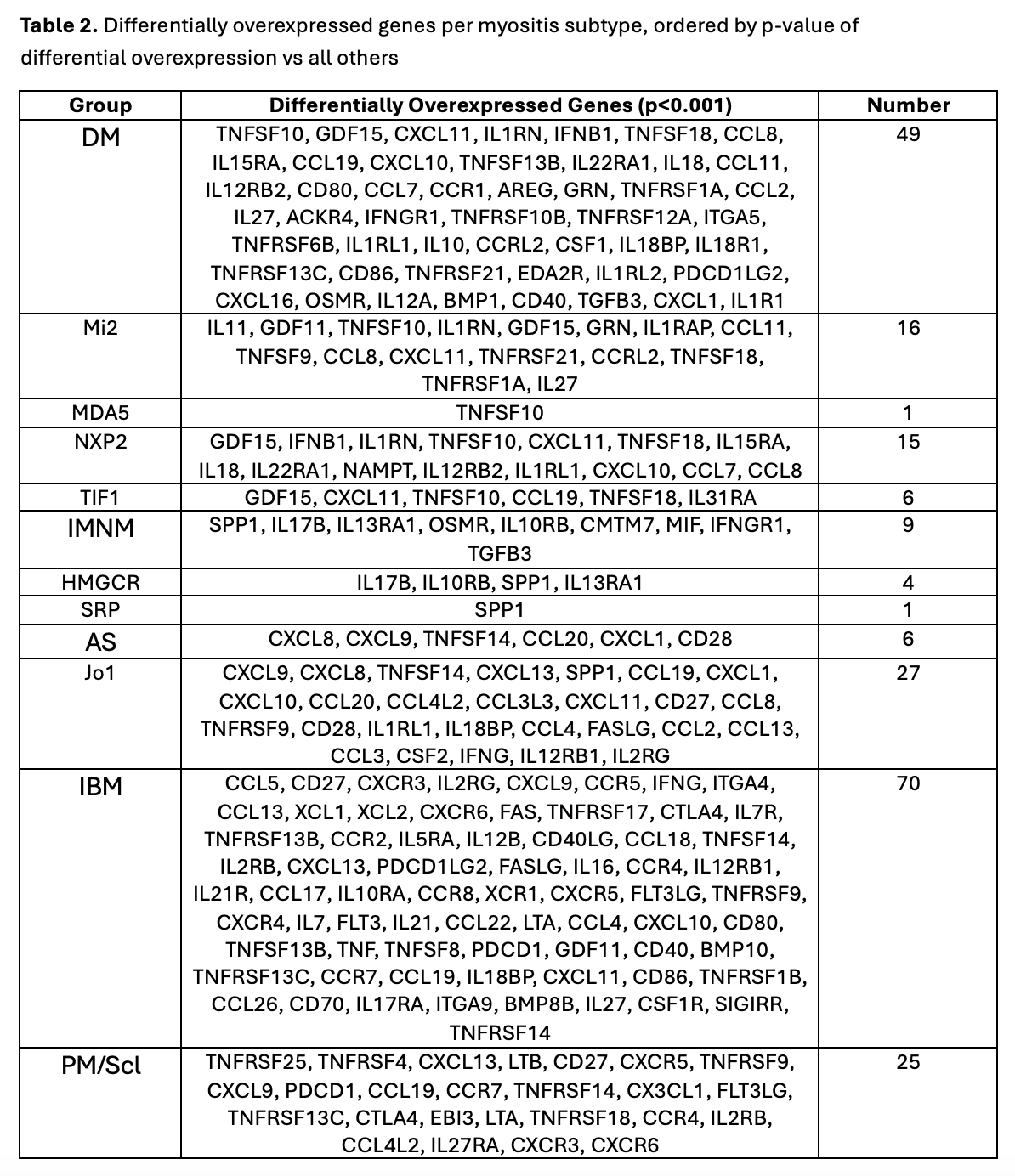
Methods: RNA-seq was performed on muscle biopsy samples from 669 patients, including 105 with dermatomyositis (DM), 80 with immune-mediated necrotizing myopathy (IMNM), 65 with anti-synthetase syndrome (AS), 53 with inclusion body myositis (IBM), and 19 with anti-PM/Scl overlap myositis (PM/Scl), as well as 310 with other inflammatory or genetic myopathies, and 37 healthy controls (NT). Myositis groups of interest were also subdivided by MSAs. RNA-seq expression data was analyzed for a list of 339 genes encoding cytokines (202), cytokine receptors (130), and checkpoint genes (7). Myositis subtype-specific genes were identified from this list by finding genes that were differentially expressed in each subtype compared to all other samples and compared to NT (α< 0.001).
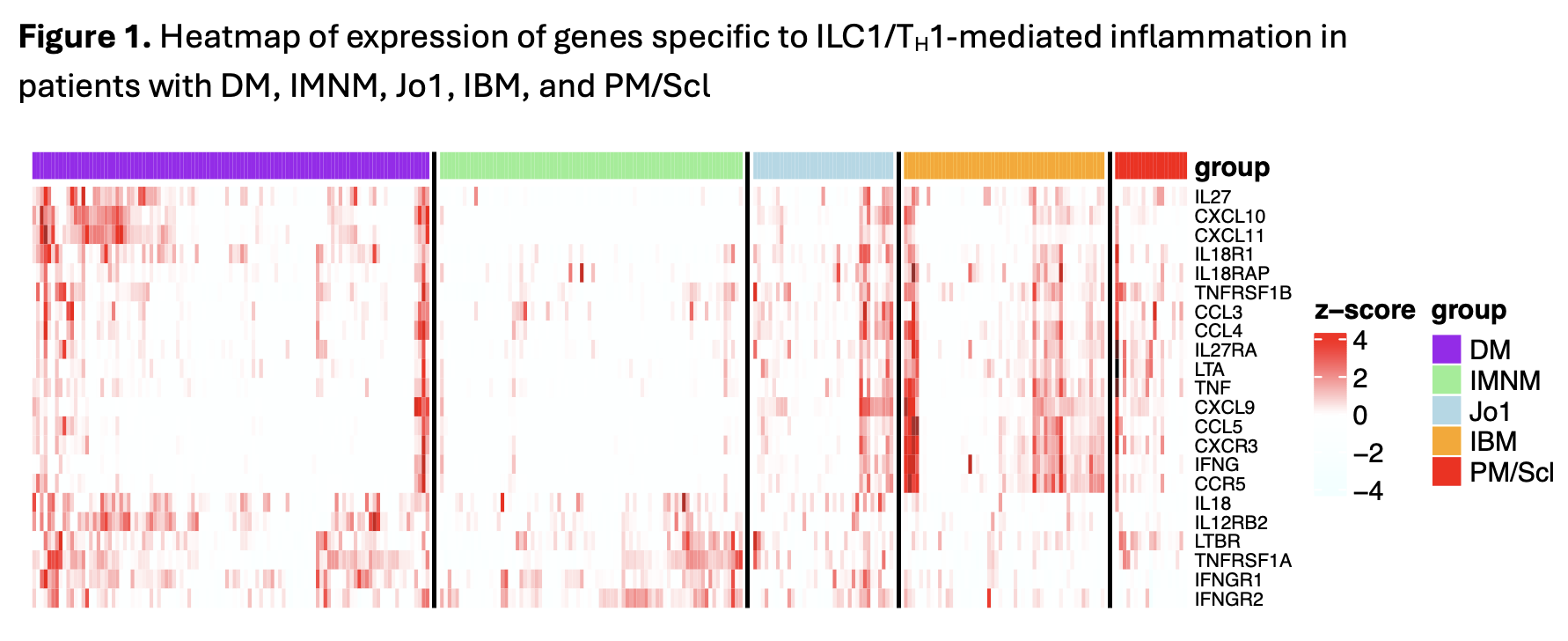
Results: Clinical groups and antibody groups analyzed are shown in Table 1. Differentially overexpressed genes for each subtype are shown in Table 2. IBM patients had the most differentially overexpressed genes (70) among all subtypes. The top 10 genes in IBM were notable for several TH1-associated genes, including the TH1 markers CXCR3 and CCR5 (and its ligand CCL5), IFNG, and the IFNG-inducible chemokine CXCL9. TH1-associated genes in IBM were strongly correlated with markers of muscle regeneration and markers of T cells, macrophages, and dendritic cells. IBM patients also displayed differential overexpression of the dendritic cell marker XCR1, and both its ligands, XCL1 and XCL2, underlying the importance of dendritic cells in promoting inflammation in IBM. Among the other groups, anti-Jo1 and anti-PM/Scl patients also exhibited overexpression (to a lesser extent) of a similar set of TH1-associated genes, while DM patients exhibited overexpression of a different set of TH1-associated genes, including CXCL10, CXCL11, and IL12RB2 (Figure 1).
Conclusion: Our analysis reveals inflammatory mediators distinctly overexpressed in different myositis subtypes, as well as others with shared overexpression between subtypes. Although some myositis subtypes are associated with autoantibodies that are thought to exert pathogenic effects on their target proteins, the pathophysiology of IBM is less understood. Our findings add support to evidence of predominantly TH1-mediated inflammation in IBM, with key involvement of IFN-γ, the CCL5-CCR5 axis, and dendritic cells. Our findings for IBM and for other autoimmune myopathies reveal key mediators of inflammation that could serve as targets for targeted therapeutic agents in myositis.
To cite this abstract in AMA style:
Kirou R, Pinal-Fernandez I, Mammen A. Distinct Cytokine and Cytokine Receptor Expression Patterns Characterize Different Subtypes of Inflammatory Myopathies [abstract]. Arthritis Rheumatol. 2024; 76 (suppl 9). https://acrabstracts.org/abstract/distinct-cytokine-and-cytokine-receptor-expression-patterns-characterize-different-subtypes-of-inflammatory-myopathies/. Accessed .« Back to ACR Convergence 2024
ACR Meeting Abstracts - https://acrabstracts.org/abstract/distinct-cytokine-and-cytokine-receptor-expression-patterns-characterize-different-subtypes-of-inflammatory-myopathies/