Session Information
Date: Monday, November 18, 2024
Title: Abstracts: Osteoporosis & Metabolic Bone Disease – Basic & Clinical Science
Session Type: Abstract Session
Session Time: 1:00PM-2:30PM
Background/Purpose: Osteonecrosis of the femoral head (ONFH) is linked to diverse risk factors like glucocorticoid use and excessive alcohol consumption. Various cells may influence bone remodeling, intraosseous pressure, and inflammation, leading to insufficient blood supply and bone tissue loss. However, the roles and interactions of bone cells in ONFH’s development and progression are not well studied. We performed bulk RNA-sequencing on ONFH femoral heads and analyzed it with public single-cell RNA-sequencing data to understand each cell type’s function in different ONFH regions.
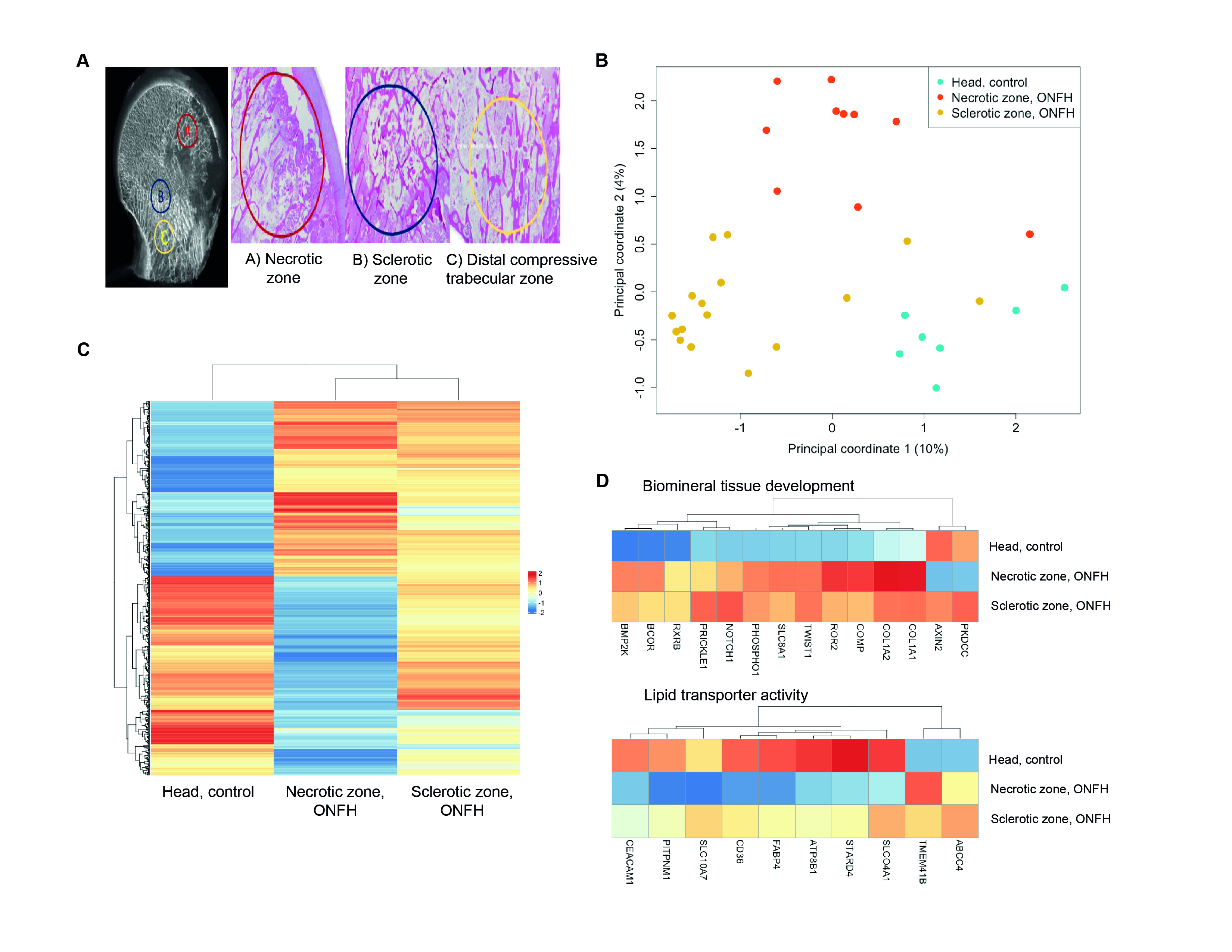
Methods: Tissue samples were collected from 17 patients with alcohol-induced (N = 5), steroid-induced (N = 8), or idiopathic (N = 4) ONFH in necrotic and sclerotic zones (Figure 1A). Control samples came from the femoral head and neck of patients with hip osteoarthritis (HOA, N = 5) or femoral neck fractures (FNF, N = 2). RNA was extracted for transcriptome analysis via bulk RNA-seq. Analyses included principal component analysis (PCA), differential expression (DE), and gene ontology (GO) enrichment using R packages edgeR, limma, and ClusterProfiler. We also analyzed public single-cell RNA-seq data from alcohol-induced ONFH patients using Seurat, excluding lymphocytes for clarity [1].
Results: We analyzed 37 samples (26 ONFH, 11 controls), identifying differentially expressed genes (DEGs). PCA on 2315 DEGs showed three well-separated groups: necrotic zone, sclerotic zone, and control head tissue (Figure 1B). Among the 2315 DEGs, 658 were differentially expressed in ONFH necrotic zones versus healthy controls, as shown in a normalized expression heatmap (Figure 1C). GO and pathway analysis highlighted two functional gene groups: ‘Biomineral tissue development’ and ‘Lipid transporter activity’ (Figure 1D).
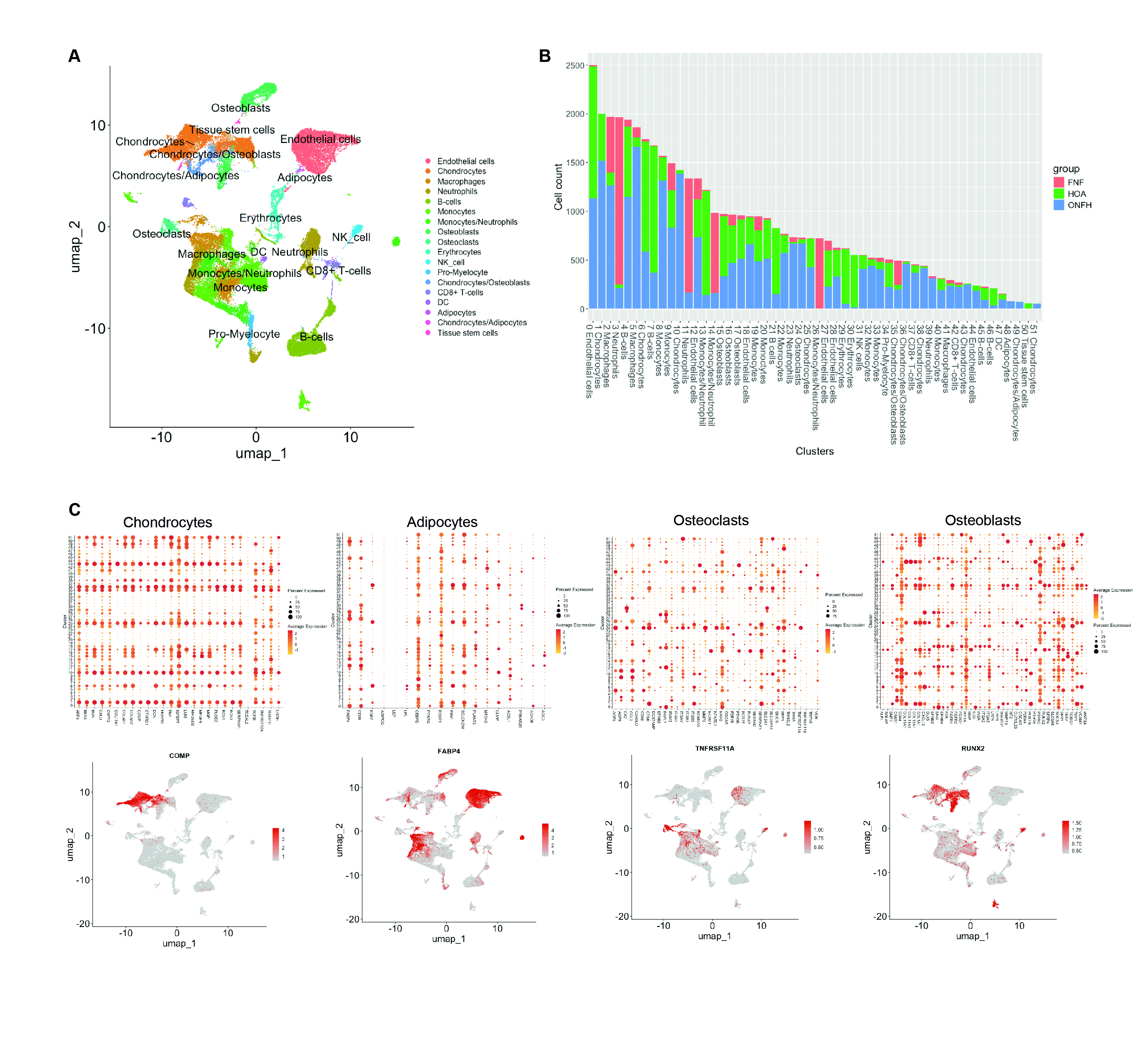
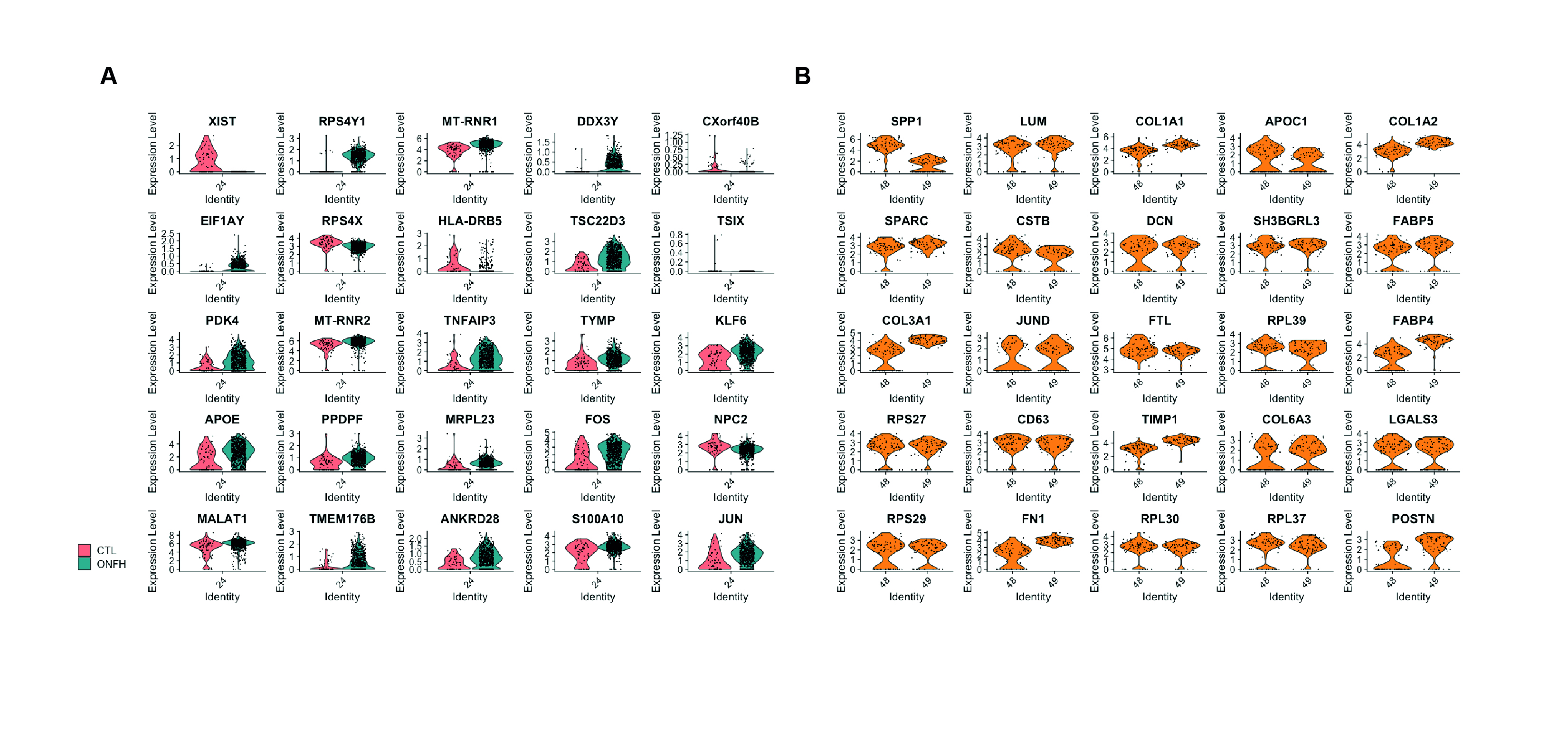
To verify bulk RNA-seq findings and identify responsible cell types, we analyzed public scRNA-seq data on alcohol-induced ONFH, HOA, and FNF. Analyzing 44,296 cells from ~80k total, we identified 51 cell clusters, including chondrocytes, adipocytes, osteoclasts, and osteoblasts (Figure 2A). Group composition in clusters varied (Figure 2B), with cell type-specific markers localized in adjacent clusters (Figure 2C). Relevant cell types from bulk RNA-seq data were examined, showing differential expression in osteoclasts and adipocytes (Figures 3A and 3B). Genes like RPS4Y1, MT-RNR1, PDK4, TNFAIP3, APOE, FOS, TMEM176B, and JUN were differentially expressed in ONFH osteoclasts. Adipocytes in ONFH increased in number, expressing SPP1, COL1A1, APOC1, CSTB, DCN, FABP5, FABP4, JUND, CD63, and TIMP1 more than other cells.
Conclusion: Our results suggest that extracellular matrix biosynthesis and lipid metabolism are key pathways in ONFH pathophysiology. Gene expression changes are differently regulated across anatomical regions and cell types.
References
1. Liao, Z. et al. Single-cell transcriptome analysis reveals aberrant stromal cells and heterogeneous endothelial cells in alcohol-induced osteonecrosis of the femoral head. Commun. Biol. 5, 324 (2022). https://doi.org/10.1038/s42003-022-03271-6
To cite this abstract in AMA style:
Lee L, Lee S, Kim M, Kim J, Shim J, Shin E, Lee T, Kwon H, Lee Y, Yang H, Park K, Song J. Gene Expression in Osteonecrosis of the Femoral Head: A Region-matched RNA-sequencing Study [abstract]. Arthritis Rheumatol. 2024; 76 (suppl 9). https://acrabstracts.org/abstract/gene-expression-in-osteonecrosis-of-the-femoral-head-a-region-matched-rna-sequencing-study/. Accessed .« Back to ACR Convergence 2024
ACR Meeting Abstracts - https://acrabstracts.org/abstract/gene-expression-in-osteonecrosis-of-the-femoral-head-a-region-matched-rna-sequencing-study/