Session Information
Session Type: Poster Session C
Session Time: 10:30AM-12:30PM
Background/Purpose: Molecular expression patterns of lung tissue across rheumatic musculoskeletal diseases (RMDs) have not been extensively studied. Lung sampling is invasive and not always practical in clinical practice. Therefore, the identification of other biological materials mirroring processes in the lung is important. The united airway disease hypothesis suggests that molecular pathways are shared between the upper and lower respiratory tracts. We determined if an altered nasal transcriptome associates with RMD-ILD; and if the nasal transcriptome is altered in subtypes of RMD-ILD and associates with progressive ILD.
Methods: Nasal mucosa samples were collected via the Copan Flocked Swab (Copan Diagnostics 56380CS01) at baseline from patients with RMD-ILD in our prospective RMD-ILD cohort study at the Oslo University Hospital and from healthy controls from UCLA and Oslo. Clinical data were collected at baseline and at 52 weeks for all RMD-ILD patients. Progressive ILD was defined as a composite of either absolute FVC% predicted decline ≥5% or DLCO% predicted decline of ≥10% and either worsening of respiratory symptoms or worsening of ILD on HRCT over 52 weeks. We assessed the transcriptome of nasal mucosa samples to identify molecular markers of RMD-ILD and compared these to healthy controls. In a second step, we assessed the samples from progressive ILD patients. The nasal transcriptome was determined by NanoString technology, using the Autoimmune Profiling panel. Gene expression and pathway analyses were conducted using ROSALIND software (https://www.rosalind.bio/).
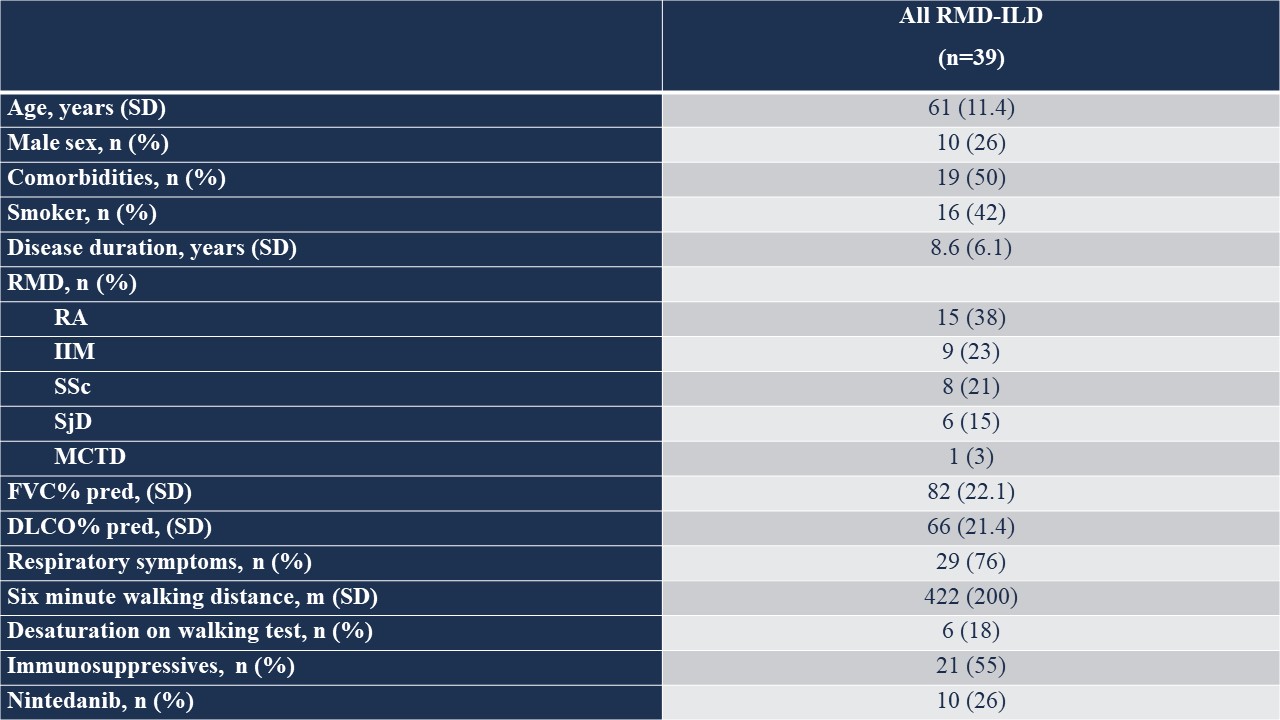
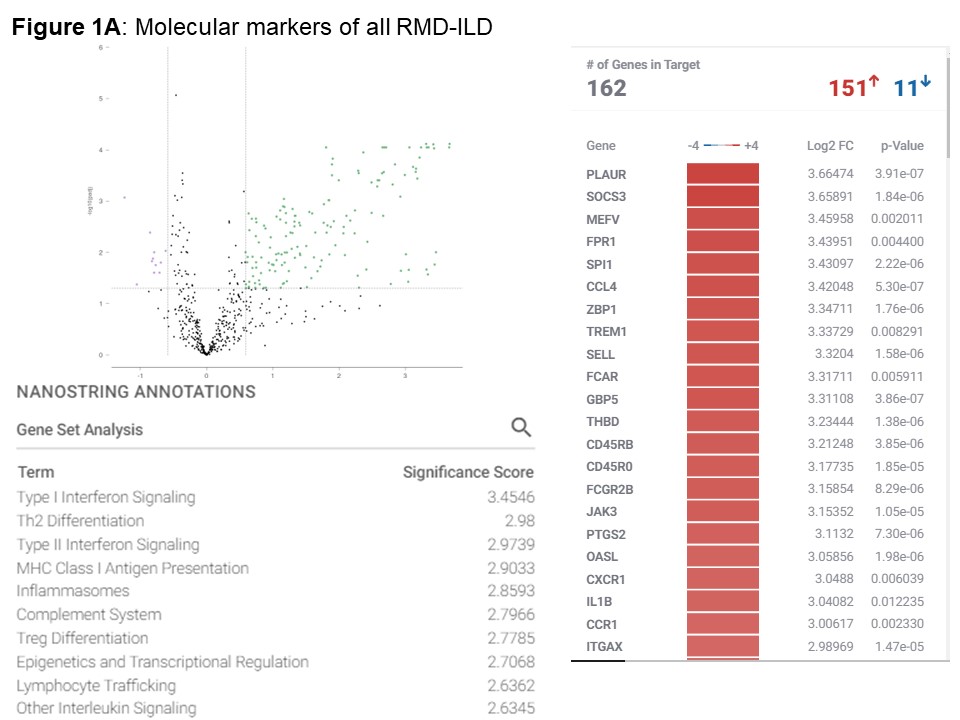
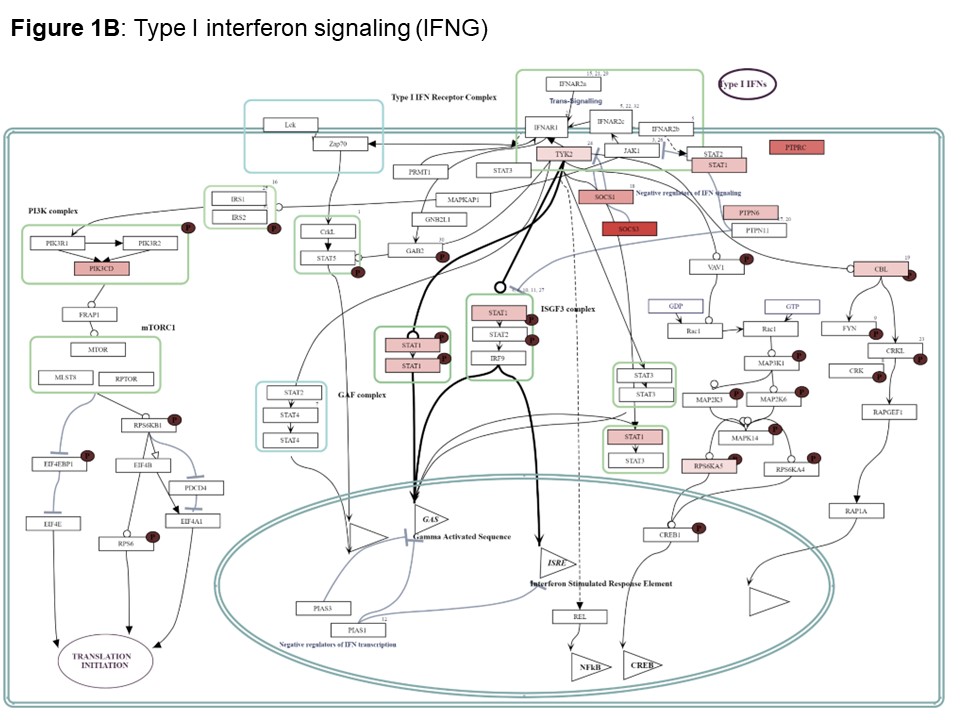
Results: We included 39 nasal mucosa samples from 15 rheumatoid arthritis (RA)-ILD, 9 idiopathic inflammatory myositis (IIM)-ILD, 8 primary Sjögren disease (SjD)-ILD, 7 systemic sclerosis (SSc)-ILD, and 1 mixed connective tissue disease (MCTD) patients and 10 healthy controls (Table). We identified 162 significant differentially expressed genes (|log2FC| ≥ 0.58 (up-regulated) or ≤ -0.58 (down-regulated), corresponding to fold change of 1.5, and adjusted p-value < 0.05) and several differentially activated signaling pathways between RMD-ILD patients and healthy controls (Figure a and 2). Several deregulated pathways were similar across the RMDs and previously identified in studies assessing RNA in lung tissue. More specifically, all single diseases were enriched for IFN-type I (all log2FC| > 1.5-3.9); and IIM, RA and SjD-ILD for TH2 differentiation (log2FC| 1.9- > 2.6). Over 52 weeks, 10 (26%) patients showed progressive ILD (5 RA-ILD, and 3 SSc, 1 SjD and 1 IIM-ILD patient). We did not identify any significantly differentially expressed genes in the progressive patients compared to the non-progressive patients.
Conclusion: We identified specific molecular markers for RMD-ILD using nasal transcriptomics from mucosa samples, which were also previously identified in studies using RNAseq on lung tissues. This justifies evaluation in further studies with larger sample sizes, as proof of concept looks promising.
To cite this abstract in AMA style:
Hoffmann-Vold A, Palchevskiy V, Diep P, Didriksen H, Talaro Ramsli M, Zhou J, Lari S, Pachera E, Durheim M, Molberg Ø, Distler O, Weigt S, Belperio J. High-throughput Screening Identifies Specific Molecular Markers for ILD in Patients with Rheumatic Diseases [abstract]. Arthritis Rheumatol. 2024; 76 (suppl 9). https://acrabstracts.org/abstract/high-throughput-screening-identifies-specific-molecular-markers-for-ild-in-patients-with-rheumatic-diseases/. Accessed .« Back to ACR Convergence 2024
ACR Meeting Abstracts - https://acrabstracts.org/abstract/high-throughput-screening-identifies-specific-molecular-markers-for-ild-in-patients-with-rheumatic-diseases/