Session Information
Date: Monday, November 18, 2024
Title: Systemic Sclerosis & Related Disorders – Basic Science Poster II
Session Type: Poster Session C
Session Time: 10:30AM-12:30PM
Background/Purpose: Lysophosphatidic Acid (LPA) is a lipid mediator implicated in the pathogenesis of SSc and idiopathic pulmonary fibrosis. Phase 2 clinical trials targeting this biological pathway show potential benefit with attenuation of downstream antifibrotic pathways through inhibition of LPA receptor 1 (LPAR1) or autotaxin. We have performed gene set enrichment analysis of skin biopsies from well characterised systemic sclerosis (SSc) patients and healthy controls using a validated 61 gene set published by the Broad Institute [https://www.gsea-msigdb.org/gsea/msigdb/index.jsp].
Methods: The BIOPSY cohort recruited 68 well characterised SSc patients prospectively from a large tertiary centre over the course of 24 months. 50 SSc skin biopsy samples were collected across disease subsets including early diffuse cutaneous (dc)SSc (n=21), limited cutaneous (lc)SSc (n=15) and established dcSSc (n=14) with 16 matched healthy controls. Bulk RNA sequencing was used to perform analyses including genome-wide transcriptome profiling of whole blood and whole skin samples. Statistical analyses was performed using Morpheus, Microsoft Excel, GraphPad Prism and STRING software packages.
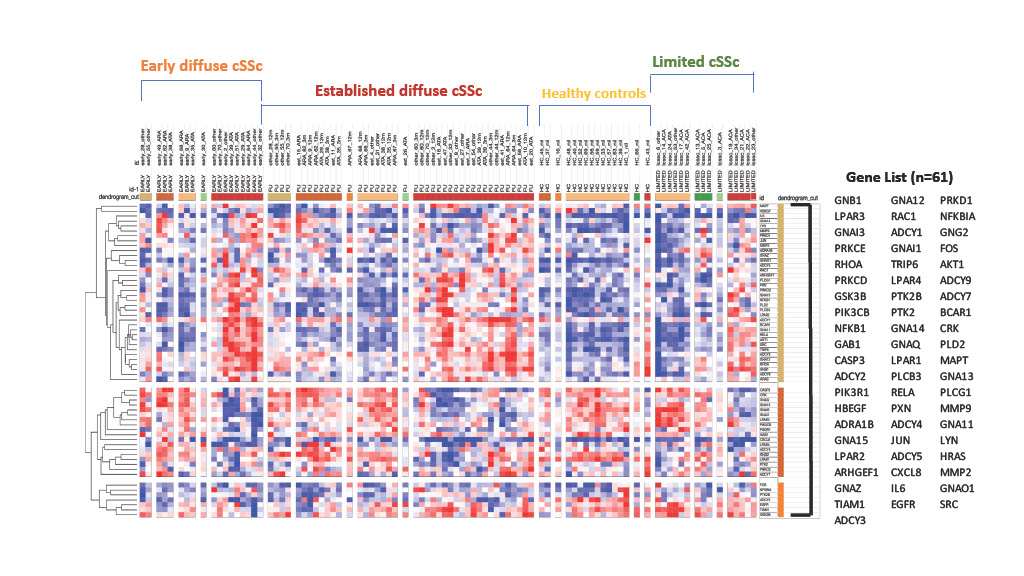
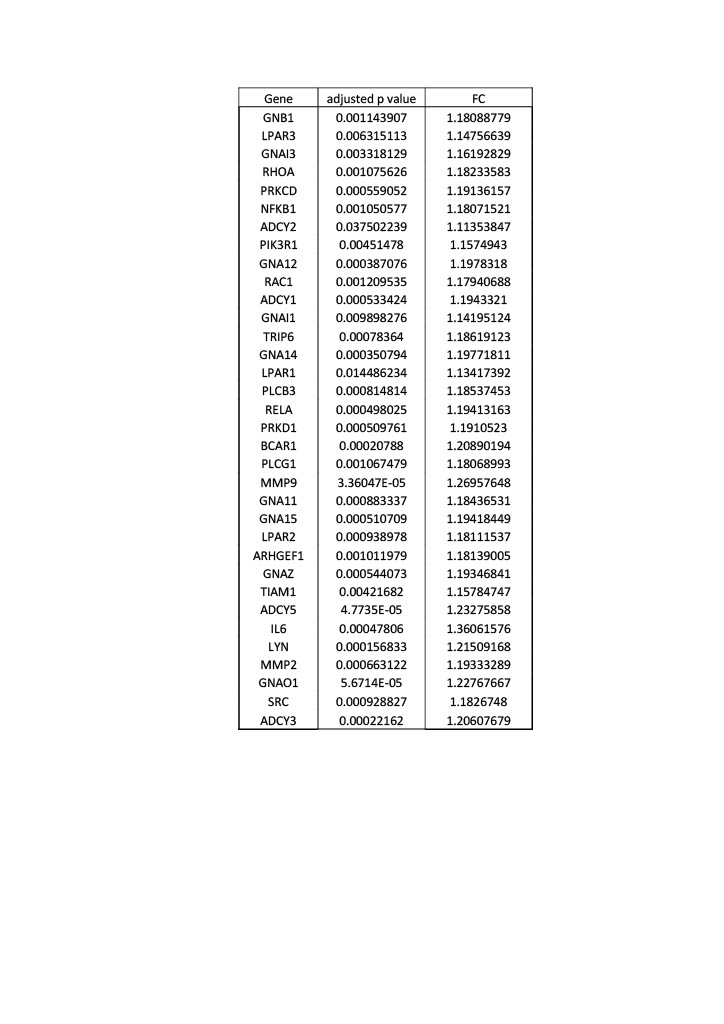
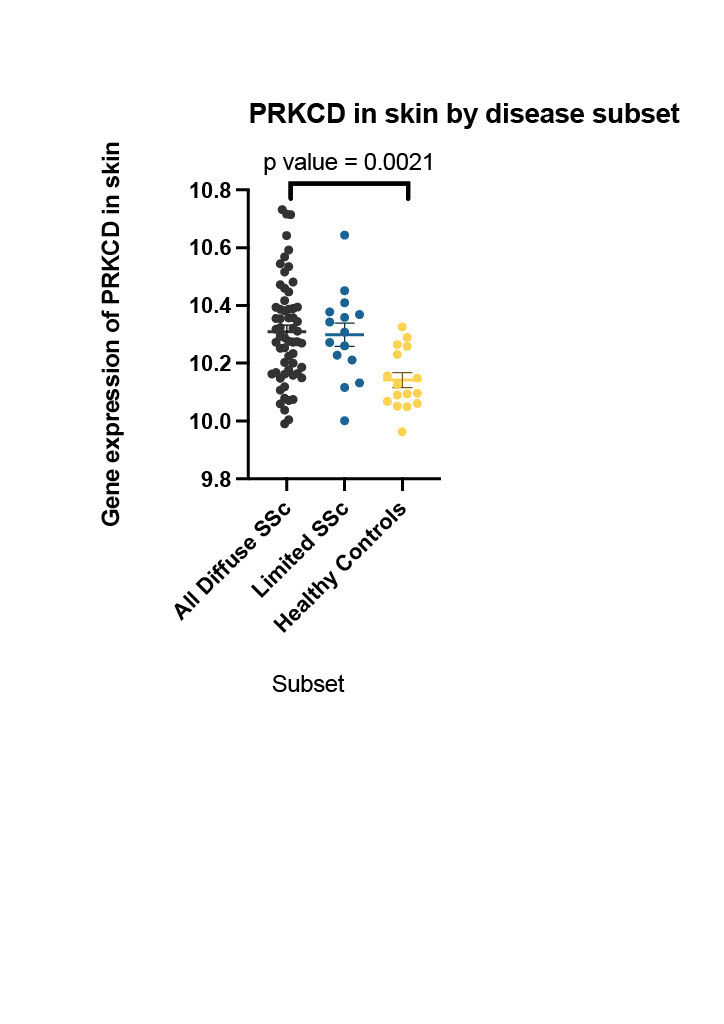
Results: Differential gene expression analysis of the published LPA gene set was used as a reference to interrogate the BIOPSY cohort subsets of SSc compared to healthy controls. Top differentially expressed LPA regulated genes by statistical significance (p value < 0.05) were identified for each SSc subset. Supervised hierarchical clustering analysis revealed distinct gene signatures across the subsets of SSc (figure 1). Unpaired t tests using adjusted p value < 0.05 applied to each SSc subset revealed differential expression between dcSSc, lcSSc and healthy controls (table 1) of certain LPA regulated genes, e.g. PRKCD shown in figure 2.
Functional analysis revealed biological pathways including D5 Dopamine receptor binding (GNA12, GNA13), calcium-independent protein kinase C activity (PRKCD and PRKCE), LPA receptor activity (LPAR1, LPAR2, LPAR3, LPAR4) and G protein-coupled serotonin receptor binding (GNAO1, GNAZ, GNA11, GNA13). 4 genes (ADCY2, LPAR1, LYN and GNAO1) with greatest statistical significance were found across the SSc disease spectrum (early dcSSc, established dcSSc and lcSSc) suggesting novel targets for future therapies.
Conclusion: Our findings confirm differential activation of LPA regulated genes in early dcSSc, established dcSSc and lcSSc that are not present in healthy skin. There is significant heterogeneity across the SSc subsets in the downstream biological pathways mediated by LPA genes in SSc. Our findings support therapeutic targeting of this pathway in SSc and identify subgroups. This may inform future clinical study design and help understand differential treatment response.
To cite this abstract in AMA style:
Kanitkar M, Yee P, Rodolfi S, Clark K, Ong V, Denton C. Analysis of Differential Activation of Lysophosphatidic Acid Regulated Genes in Diffuse and Limited Cutaneous Systemic Sclerosis [abstract]. Arthritis Rheumatol. 2024; 76 (suppl 9). https://acrabstracts.org/abstract/analysis-of-differential-activation-of-lysophosphatidic-acid-regulated-genes-in-diffuse-and-limited-cutaneous-systemic-sclerosis/. Accessed .« Back to ACR Convergence 2024
ACR Meeting Abstracts - https://acrabstracts.org/abstract/analysis-of-differential-activation-of-lysophosphatidic-acid-regulated-genes-in-diffuse-and-limited-cutaneous-systemic-sclerosis/